A new method to extract dental pulp DNA: application to universal detection of bacteria
- PMID: 17957246
- PMCID: PMC2031827
- DOI: 10.1371/journal.pone.0001062
A new method to extract dental pulp DNA: application to universal detection of bacteria
Abstract
Background: Dental pulp is used for PCR-based detection of DNA derived from host and bacteremic microorganims. Current protocols require odontology expertise for proper recovery of the dental pulp. Dental pulp specimen exposed to laboratory environment yields contaminants detected using universal 16S rDNA-based detection of bacteria.
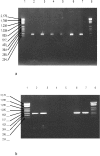
Methodology/principal findings: We developed a new protocol by encasing decontaminated tooth into sterile resin, extracting DNA into the dental pulp chamber itself and decontaminating PCR reagents by filtration and double restriction enzyme digestion. Application to 16S rDNA-based detection of bacteria in 144 teeth collected in 86 healthy people yielded a unique sequence in only 14 teeth (9.7%) from 12 individuals (14%). Each individual yielded a unique 16S rDNA sequence in 1-2 teeth per individual. Negative controls remained negative. Bacterial identifications were all confirmed by amplification and sequencing of specific rpoB sequence.
Conclusions/significance: The new protocol prevented laboratory contamination of the dental pulp. It allowed the detection of bacteria responsible for dental pulp colonization from blood and periodontal tissue. Only 10% such samples contained 16S rDNA. It provides a new tool for the retrospective diagnostic of bacteremia by allowing the universal detection of bacterial DNA in animal and human, contemporary or ancient tooth. It could be further applied to identification of host DNA in forensic medicine and anthropology.
Conflict of interest statement
Figures


Similar articles
-
Detection of 400-year-old Yersinia pestis DNA in human dental pulp: an approach to the diagnosis of ancient septicemia.Proc Natl Acad Sci U S A. 1998 Oct 13;95(21):12637-40. doi: 10.1073/pnas.95.21.12637. Proc Natl Acad Sci U S A. 1998. PMID: 9770538 Free PMC article.
-
Detection of bacteremia in emergency department patients at risk for infective endocarditis using universal 16S rRNA primers in a decontaminated polymerase chain reaction assay.J Infect Dis. 2002 Dec 1;186(11):1677-81. doi: 10.1086/345367. Epub 2002 Nov 6. J Infect Dis. 2002. PMID: 12447747
-
Molecular detection of Bartonella quintana DNA in the dental pulp of a homeless patient.Eur J Clin Microbiol Infect Dis. 2004 Dec;23(12):920-2. doi: 10.1007/s10096-004-1244-z. Eur J Clin Microbiol Infect Dis. 2004. PMID: 15558347
-
Review and re-analysis of domain-specific 16S primers.J Microbiol Methods. 2003 Dec;55(3):541-55. doi: 10.1016/j.mimet.2003.08.009. J Microbiol Methods. 2003. PMID: 14607398 Review.
-
Ancient dental pulp: Masterpiece tissue for paleomicrobiology.Mol Genet Genomic Med. 2020 Jun;8(6):e1202. doi: 10.1002/mgg3.1202. Epub 2020 Mar 31. Mol Genet Genomic Med. 2020. PMID: 32233019 Free PMC article. Review.
Cited by
-
Using comparative genomics for inquiry-based learning to dissect virulence of Escherichia coli O157:H7 and Yersinia pestis.CBE Life Sci Educ. 2012 Spring;11(1):81-93. doi: 10.1187/cbe.10-04-0057. CBE Life Sci Educ. 2012. PMID: 22383620 Free PMC article.
-
Decontamination by Persteril 36 may affect the reliability of DNA-based detection of biological warfare agents-short communication.Folia Microbiol (Praha). 2016 Sep;61(5):417-21. doi: 10.1007/s12223-016-0451-1. Epub 2016 Feb 24. Folia Microbiol (Praha). 2016. PMID: 26910525
-
Detection of HCV Persistent Infections in the Dental Pulp: A Novel Approach for the Detection of Past and Ancient Infections.PLoS One. 2016 Oct 26;11(10):e0165272. doi: 10.1371/journal.pone.0165272. eCollection 2016. PLoS One. 2016. PMID: 27783693 Free PMC article.
-
Paleoproteomics of the Dental Pulp: The plague paradigm.PLoS One. 2017 Jul 26;12(7):e0180552. doi: 10.1371/journal.pone.0180552. eCollection 2017. PLoS One. 2017. PMID: 28746380 Free PMC article.
-
Tracing back ancient oral microbiomes and oral pathogens using dental pulps from ancient teeth.NPJ Biofilms Microbiomes. 2016 Dec 7;2:6. doi: 10.1038/s41522-016-0008-8. eCollection 2016. NPJ Biofilms Microbiomes. 2016. PMID: 28649400 Free PMC article.
References
-
- Delivanis PD, Snowden RB, Doyle RJ. Localization of blood-borne bacteria in instrumented unfilled root canals. Oral Surg Oral Med Oral Pathol. 1981 Oct;52(4):430–2. - PubMed
-
- Tziafas D. Experimental bacterial anachoresis in dog dental pulps capped with calcium hydroxide. J Endod. 1989 Dec;15(12):591–5. - PubMed
-
- Hoshino E, Ando N, Sato M, Kota K. Bacterial invasion of non-exposed dental pulp. Int Endod J. 1992 Jan;25(1):2–5. - PubMed
-
- Aboudharam G, La Scola B, Raoult D, Drancourt M. Detection of Coxiella burnetii DNA in dental pulp during experimental bacteremia. Microb Pathog. 2000;28(4):249–54. - PubMed
-
- Aboudharam G, Drancourt M, Raoult D. Culture of C. burnetii from the dental pulp of experimentally infected guinea pigs. Microb Pathog. 2004 Jun;36(6):349–50. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

