Microarray analysis in the archaeon Halobacterium salinarum strain R1
- PMID: 17957248
- PMCID: PMC2020435
- DOI: 10.1371/journal.pone.0001064
Microarray analysis in the archaeon Halobacterium salinarum strain R1
Abstract
Background: Phototrophy of the extremely halophilic archaeon Halobacterium salinarum was explored for decades. The research was mainly focused on the expression of bacteriorhodopsin and its functional properties. In contrast, less is known about genome wide transcriptional changes and their impact on the physiological adaptation to phototrophy. The tool of choice to record transcriptional profiles is the DNA microarray technique. However, the technique is still rarely used for transcriptome analysis in archaea.
Methodology/principal findings: We developed a whole-genome DNA microarray based on our sequence data of the Hbt. salinarum strain R1 genome. The potential of our tool is exemplified by the comparison of cells growing under aerobic and phototrophic conditions, respectively. We processed the raw fluorescence data by several stringent filtering steps and a subsequent MAANOVA analysis. The study revealed a lot of transcriptional differences between the two cell states. We found that the transcriptional changes were relatively weak, though significant. Finally, the DNA microarray data were independently verified by a real-time PCR analysis.
Conclusion/significance: This is the first DNA microarray analysis of Hbt. salinarum cells that were actually grown under phototrophic conditions. By comparing the transcriptomics data with current knowledge we could show that our DNA microarray tool is well applicable for transcriptome analysis in the extremely halophilic archaeon Hbt. salinarum. The reliability of our tool is based on both the high-quality array of DNA probes and the stringent data handling including MAANOVA analysis. Among the regulated genes more than 50% had unknown functions. This underlines the fact that haloarchaeal phototrophy is still far away from being completely understood. Hence, the data recorded in this study will be subject to future systems biology analysis.
Conflict of interest statement
Figures



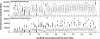

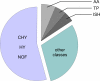
Similar articles
-
Variations in the multiple tbp genes in different Halobacterium salinarum strains and their expression during growth.Arch Microbiol. 2008 Sep;190(3):309-18. doi: 10.1007/s00203-008-0383-5. Epub 2008 May 28. Arch Microbiol. 2008. PMID: 18506423
-
Genome-wide analysis of growth phase-dependent translational and transcriptional regulation in halophilic archaea.BMC Genomics. 2007 Nov 12;8:415. doi: 10.1186/1471-2164-8-415. BMC Genomics. 2007. PMID: 17997854 Free PMC article.
-
Archaeal transcriptional regulation of the prokaryotic KdpFABC complex mediating K(+) uptake in H. salinarum.Extremophiles. 2011 Nov;15(6):643-52. doi: 10.1007/s00792-011-0395-y. Epub 2011 Sep 21. Extremophiles. 2011. PMID: 21947979
-
Global transcriptional regulator TrmB family members in prokaryotes.J Microbiol. 2016 Oct;54(10):639-45. doi: 10.1007/s12275-016-6362-7. Epub 2016 Sep 30. J Microbiol. 2016. PMID: 27687225 Review.
-
Global Transcriptional Programs in Archaea Share Features with the Eukaryotic Environmental Stress Response.J Mol Biol. 2019 Sep 20;431(20):4147-4166. doi: 10.1016/j.jmb.2019.07.029. Epub 2019 Aug 19. J Mol Biol. 2019. PMID: 31437442 Free PMC article. Review.
Cited by
-
Phosphate-dependent behavior of the archaeon Halobacterium salinarum strain R1.J Bacteriol. 2009 Jun;191(12):3852-60. doi: 10.1128/JB.01642-08. Epub 2009 Apr 10. J Bacteriol. 2009. PMID: 19363117 Free PMC article.
-
Towards a systems approach in the genetic analysis of archaea: Accelerating mutant construction and phenotypic analysis in Haloferax volcanii.Archaea. 2010 Dec 23;2010:426239. doi: 10.1155/2010/426239. Archaea. 2010. PMID: 21234384 Free PMC article.
-
Novel pili-like surface structures of Halobacterium salinarum strain R1 are crucial for surface adhesion.Front Microbiol. 2015 Jan 13;5:755. doi: 10.3389/fmicb.2014.00755. eCollection 2014. Front Microbiol. 2015. PMID: 25628607 Free PMC article.
-
Gene Expression of Haloferax volcanii on Intermediate and Abundant Sources of Fixed Nitrogen.Int J Mol Sci. 2019 Sep 26;20(19):4784. doi: 10.3390/ijms20194784. Int J Mol Sci. 2019. PMID: 31561502 Free PMC article.
-
A small protein from the bop-brp intergenic region of Halobacterium salinarum contains a zinc finger motif and regulates bop and crtB1 transcription.Mol Microbiol. 2008 Feb;67(4):772-80. doi: 10.1111/j.1365-2958.2007.06081.x. Epub 2008 Jan 2. Mol Microbiol. 2008. PMID: 18179416 Free PMC article.
References
-
- Sumper M, Reitmeier H, Oesterhelt D. Biosynthesis of the purple membrane of halobacteria. Angew Chem Int Ed Engl. 1976;15:187–94. - PubMed
-
- Sumper M, Herrmann G. Biogenesis of purple membrane: regulation of bacterio-opsin synthesis. FEBS Lett. 1976;69:149–52. - PubMed
-
- Oesterhelt D, Krippahl G. Phototrophic growth of halobacteria and its use for isolation of photosynthetically-deficient mutants. Ann Microbiol. 1983;B134:137–150. - PubMed
-
- Peck RF, Echavarri-Erasun C, Johnson EA, Ng WV, Kennedy SP, et al. brp and blh are required for synthesis of the retinal cofactor of bacteriorhodopsin in Halobacterium salinarum. J Biol Chem. 2001;276:5739–5744. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

