Ancestral inference and the study of codon bias evolution: implications for molecular evolutionary analyses of the Drosophila melanogaster subgroup
- PMID: 17957249
- PMCID: PMC2020436
- DOI: 10.1371/journal.pone.0001065
Ancestral inference and the study of codon bias evolution: implications for molecular evolutionary analyses of the Drosophila melanogaster subgroup
Abstract
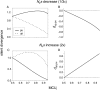
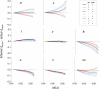
Reliable inference of ancestral sequences can be critical to identifying both patterns and causes of molecular evolution. Robustness of ancestral inference is often assumed among closely related species, but tests of this assumption have been limited. Here, we examine the performance of inference methods for data simulated under scenarios of codon bias evolution within the Drosophila melanogaster subgroup. Genome sequence data for multiple, closely related species within this subgroup make it an important system for studying molecular evolutionary genetics. The effects of asymmetric and lineage-specific substitution rates (i.e., varying levels of codon usage bias and departures from equilibrium) on the reliability of ancestral codon usage was investigated. Maximum parsimony inference, which has been widely employed in analyses of Drosophila codon bias evolution, was compared to an approach that attempts to account for uncertainty in ancestral inference by weighting ancestral reconstructions by their posterior probabilities. The latter approach employs maximum likelihood estimation of rate and base composition parameters. For equilibrium and most non-equilibrium scenarios that were investigated, the probabilistic method appears to generate reliable ancestral codon bias inferences for molecular evolutionary studies within the D. melanogaster subgroup. These reconstructions are more reliable than parsimony inference, especially when codon usage is strongly skewed. However, inference biases are considerable for both methods under particular departures from stationarity (i.e., when adaptive evolution is prevalent). Reliability of inference can be sensitive to branch lengths, asymmetry in substitution rates, and the locations and nature of lineage-specific processes within a gene tree. Inference reliability, even among closely related species, can be strongly affected by (potentially unknown) patterns of molecular evolution in lineages ancestral to those of interest.
Conflict of interest statement
Figures
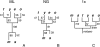



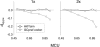


Similar articles
-
Molecular evolution in the Drosophila melanogaster species subgroup: frequent parameter fluctuations on the timescale of molecular divergence.Genetics. 2006 Mar;172(3):1711-26. doi: 10.1534/genetics.105.049676. Epub 2005 Dec 30. Genetics. 2006. PMID: 16387879 Free PMC article.
-
Codon Usage Selection Can Bias Estimation of the Fraction of Adaptive Amino Acid Fixations.Mol Biol Evol. 2016 Jun;33(6):1580-9. doi: 10.1093/molbev/msw027. Epub 2016 Feb 12. Mol Biol Evol. 2016. PMID: 26873577
-
Molecular evolution between Drosophila melanogaster and D. simulans: reduced codon bias, faster rates of amino acid substitution, and larger proteins in D. melanogaster.Genetics. 1996 Nov;144(3):1297-307. doi: 10.1093/genetics/144.3.1297. Genetics. 1996. PMID: 8913769 Free PMC article.
-
Patterns of polymorphism and divergence from noncoding sequences of Drosophila melanogaster and D. simulans: evidence for nonequilibrium processes.Mol Biol Evol. 2005 Jan;22(1):51-62. doi: 10.1093/molbev/msh269. Epub 2004 Sep 29. Mol Biol Evol. 2005. PMID: 15456897 Review.
-
Mutation pressure, natural selection, and the evolution of base composition in Drosophila.Genetica. 1998;102-103(1-6):49-60. Genetica. 1998. PMID: 9720271 Review.
Cited by
-
Accelerated sequence divergence of conserved genomic elements in Drosophila melanogaster.Genome Res. 2008 Oct;18(10):1592-601. doi: 10.1101/gr.077131.108. Epub 2008 Jun 26. Genome Res. 2008. PMID: 18583644 Free PMC article.
-
Distinguishing Among Evolutionary Forces Acting on Genome-Wide Base Composition: Computer Simulation Analysis of Approximate Methods for Inferring Site Frequency Spectra of Derived Mutations.G3 (Bethesda). 2018 May 4;8(5):1755-1769. doi: 10.1534/g3.117.300512. G3 (Bethesda). 2018. PMID: 29588382 Free PMC article.
-
Locus-specific decoupling of base composition evolution at synonymous sites and introns along the Drosophila melanogaster and Drosophila sechellia lineages.Genome Biol Evol. 2009 May 25;1:67-74. doi: 10.1093/gbe/evp008. Genome Biol Evol. 2009. PMID: 20333178 Free PMC article.
-
Population genomic analysis of base composition evolution in Drosophila melanogaster.Genome Biol Evol. 2012;4(12):1245-55. doi: 10.1093/gbe/evs097. Genome Biol Evol. 2012. PMID: 23160062 Free PMC article.
-
Optimized ancestral state reconstruction using Sankoff parsimony.BMC Bioinformatics. 2009 Feb 7;10:51. doi: 10.1186/1471-2105-10-51. BMC Bioinformatics. 2009. PMID: 19200389 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

