Conformational reorganization of the SARS coronavirus spike following receptor binding: implications for membrane fusion
- PMID: 17957264
- PMCID: PMC2034598
- DOI: 10.1371/journal.pone.0001082
Conformational reorganization of the SARS coronavirus spike following receptor binding: implications for membrane fusion
Abstract
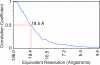
The SARS coronavirus (SARS-CoV) spike is the largest known viral spike molecule, and shares a similar function with all class 1 viral fusion proteins. Previous structural studies of membrane fusion proteins have largely used crystallography of static molecular fragments, in isolation of their transmembrane domains. In this study we have produced purified, irradiated SARS-CoV virions that retain their morphology, and are fusogenic in cell culture. We used cryo-electron microscopy and image processing to investigate conformational changes that occur in the entire spike of intact virions when they bind to the viral receptor, angiotensin-converting enzyme 2 (ACE2). We have shown that ACE2 binding results in structural changes that appear to be the initial step in viral membrane fusion, and precisely localized the receptor-binding and fusion core domains within the entire spike. Furthermore, our results show that receptor binding and subsequent membrane fusion are distinct steps, and that each spike can bind up to three ACE2 molecules. The SARS-CoV spike provides an ideal model system to study receptor binding and membrane fusion in the native state, employing cryo-electron microscopy and single-particle image analysis.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

