A nucleosome positioned by alpha2/Mcm1 prevents Hap1 activator binding in vivo
- PMID: 17959145
- PMCID: PMC2131697
- DOI: 10.1016/j.bbrc.2007.10.037
A nucleosome positioned by alpha2/Mcm1 prevents Hap1 activator binding in vivo
Abstract
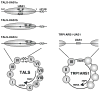
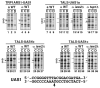
Nucleosome positioning has been proposed as a mechanism of transcriptional repression. Here, we examined whether nucleosome positioning affects activator binding in living yeast cells. We introduced the cognate Hap1 binding site (UAS1) at a location 24-43 bp, 29-48 bp, or 61-80 bp interior to the edge of a nucleosome positioned by alpha2/Mcm1 in yeast minichromosomes. Hap1 binding to the UAS1 was severely inhibited, not only at the pseudo-dyad but also in the peripheral region of the positioned nucleosome in alpha cells, while it was detectable in a cells, in which the nucleosomes were not positioned. Hap1 binding was restored in alpha cells with tup1 or isw2 mutations, which caused the loss of nucleosome positioning. These results support the mechanism in which alpha2/Mcm1-dependent nucleosome positioning has a regulatory function to limit the access of transcription factors.
Figures

Similar articles
-
Effect of sequence-directed nucleosome disruption on cell-type-specific repression by alpha2/Mcm1 in the yeast genome.Eukaryot Cell. 2006 Nov;5(11):1925-33. doi: 10.1128/EC.00105-06. Epub 2006 Sep 15. Eukaryot Cell. 2006. PMID: 16980406 Free PMC article.
-
Roles of transcription factor Mot3 and chromatin in repression of the hypoxic gene ANB1 in yeast.Mol Cell Biol. 2000 Oct;20(19):7088-98. doi: 10.1128/MCB.20.19.7088-7098.2000. Mol Cell Biol. 2000. PMID: 10982825 Free PMC article.
-
The coiled coil dimerization element of the yeast transcriptional activator Hap1, a Gal4 family member, is dispensable for DNA binding but differentially affects transcriptional activation.J Biol Chem. 2000 Jan 7;275(1):248-54. doi: 10.1074/jbc.275.1.248. J Biol Chem. 2000. PMID: 10617612
-
Mutations that alter transcriptional activation but not DNA binding in the zinc finger of yeast activator HAPI.Nature. 1989 Nov 9;342(6246):200-3. doi: 10.1038/342200a0. Nature. 1989. PMID: 2509943
-
Nucleosome positioning--what do we really know?Mol Biosyst. 2009 Dec;5(12):1582-92. doi: 10.1039/b907227f. Epub 2009 Sep 30. Mol Biosyst. 2009. PMID: 19795076 Review.
Cited by
-
The Cyc8-Tup1 complex inhibits transcription primarily by masking the activation domain of the recruiting protein.Genes Dev. 2011 Dec 1;25(23):2525-39. doi: 10.1101/gad.179275.111. Genes Dev. 2011. PMID: 22156212 Free PMC article.
-
Telomeric repeats act as nucleosome-disfavouring sequences in vivo.Nucleic Acids Res. 2014 Feb;42(3):1541-52. doi: 10.1093/nar/gkt1006. Epub 2013 Oct 29. Nucleic Acids Res. 2014. PMID: 24174540 Free PMC article.
-
Nucleosome-coupled expression differences in closely-related species.BMC Genomics. 2011 Sep 26;12:466. doi: 10.1186/1471-2164-12-466. BMC Genomics. 2011. PMID: 21942931 Free PMC article.
-
Nucleosome-depleted regions in cell-cycle-regulated promoters ensure reliable gene expression in every cell cycle.Dev Cell. 2010 Apr 20;18(4):544-55. doi: 10.1016/j.devcel.2010.02.007. Dev Cell. 2010. PMID: 20412770 Free PMC article.
-
Role of the cell wall integrity and filamentous growth mitogen-activated protein kinase pathways in cell wall remodeling during filamentous growth.Eukaryot Cell. 2009 Aug;8(8):1118-33. doi: 10.1128/EC.00006-09. Epub 2009 Jun 5. Eukaryot Cell. 2009. PMID: 19502582 Free PMC article.
References
-
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. - PubMed
-
- Morse RH. Transcription factor access to promoter elements. J Cell Biochem. 2007 - PubMed
-
- Owen-Hughes T, Workman JL. Experimental analysis of chromatin function in transcription control. Crit Rev Eukaryot Gene Expr. 1994;4:403–441. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

