Dynamic clustered distribution of hemagglutinin resolved at 40 nm in living cell membranes discriminates between raft theories
- PMID: 17959773
- PMCID: PMC2077263
- DOI: 10.1073/pnas.0708066104
Dynamic clustered distribution of hemagglutinin resolved at 40 nm in living cell membranes discriminates between raft theories
Abstract
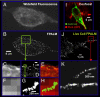
Organization in biological membranes spans many orders of magnitude in length scale, but limited resolution in far-field light microscopy has impeded distinction between numerous biomembrane models. One canonical example of a heterogeneously distributed membrane protein is hemagglutinin (HA) from influenza virus, which is associated with controversial cholesterol-rich lipid rafts. Using fluorescence photoactivation localization microscopy, we are able to image distributions of tens of thousands of HA molecules with subdiffraction resolution ( approximately 40 nm) in live and fixed fibroblasts. HA molecules form irregular clusters on length scales from approximately 40 nm up to many micrometers, consistent with results from electron microscopy. In live cells, the dynamics of HA molecules within clusters is observed and quantified to determine an effective diffusion coefficient. The results are interpreted in terms of several established models of biological membranes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

