Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast
- PMID: 17959824
- PMCID: PMC2174172
- DOI: 10.1091/mbc.e07-08-0779
Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast
Abstract
We studied the relationship between growth rate and genome-wide gene expression, cell cycle progression, and glucose metabolism in 36 steady-state continuous cultures limited by one of six different nutrients (glucose, ammonium, sulfate, phosphate, uracil, or leucine). The expression of more than one quarter of all yeast genes is linearly correlated with growth rate, independent of the limiting nutrient. The subset of negatively growth-correlated genes is most enriched for peroxisomal functions, whereas positively correlated genes mainly encode ribosomal functions. Many (not all) genes associated with stress response are strongly correlated with growth rate, as are genes that are periodically expressed under conditions of metabolic cycling. We confirmed a linear relationship between growth rate and the fraction of the cell population in the G0/G1 cell cycle phase, independent of limiting nutrient. Cultures limited by auxotrophic requirements wasted excess glucose, whereas those limited on phosphate, sulfate, or ammonia did not; this phenomenon (reminiscent of the "Warburg effect" in cancer cells) was confirmed in batch cultures. Using an aggregate of gene expression values, we predict (in both continuous and batch cultures) an "instantaneous growth rate." This concept is useful in interpreting the system-level connections among growth rate, metabolism, stress, and the cell cycle.
Figures
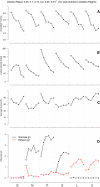

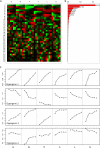

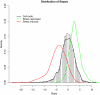
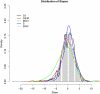
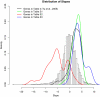
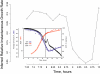
References
-
- Airoldi E. M., Huttenhower C., Gresham D., Botstein D., Troyanskaya O. G. Growth-specific programs of gene expression. Proceedings of the Problems and Methods in Computational Biology Workshop; 2007. NIPS 2007.
-
- Alexander M., Jeffries T. Respiratory efficiency and metabolite partitioning as regulatory phenomena in yeasts. Enzyme Microb. Technol. 1990;12:2–19.
-
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995;57:289–300.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

