Chromatin challenges during DNA replication: a systems representation
- PMID: 17959828
- PMCID: PMC2174177
- DOI: 10.1091/mbc.e07-06-0528
Chromatin challenges during DNA replication: a systems representation
Abstract
In a recent review, A. Groth and coworkers presented a comprehensive account of nucleosome disassembly in front of a DNA replication fork, assembly behind the replication fork, and the copying of epigenetic information onto the replicated chromatin. Understanding those processes however would be enhanced by a comprehensive graphical depiction analogous to a circuit diagram. Accordingly, we have constructed a molecular interaction map (MIM) that preserves in essentially complete detail the processes described by Groth et al. The MIM organizes and elucidates the information presented by Groth et al. on the complexities of chromatin replication, thereby providing a tool for system-level comprehension of the effects of genetic mutations, altered gene expression, and pharmacologic intervention.
Figures

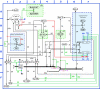
References
-
- Aladjem M. I., Pasa S., Parodi S., Weinstein J. N., Pommier Y., Kohn K. W. Molecular interaction maps—a diagrammatic graphical language for bioregulatory networks. Sci. STKE. 2004;2004:pe8. - PubMed
-
- Groth A., Rocha W., Verreault A., Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. - PubMed
ANNOTATION REFERENCES
-
- Barman H. K., Takami Y., Ono T., Nishijima H., Sanematsu F., Shibahara K., Nakayama T. Histone acetyltransferase 1 is dispensable for replication-coupled chromatin assembly but contributes to recover DNA damages created following replication blockage in vertebrate cells. Biochem. Biophys. Res. Commun. 2006;345:1547–1557. - PubMed
-
- Belotserkovskaya R., Reinberg D. Facts about FACT and transcript elongation through chromatin. Curr. Opin. Genet. Dev. 2004;14:139–146. - PubMed
-
- Chuang L. S., Ian H. I., Koh T. W., Ng H. H., Xu G., Li B. F. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

