Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone
- PMID: 17960235
- PMCID: PMC2581903
- DOI: 10.1038/nature06235
Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone
Abstract
DExD/H-box proteins are ubiquitously involved in RNA-mediated processes and use ATP to accelerate conformational changes in RNA. However, their mechanisms of action, and what determines which RNA species are targeted, are not well understood. Here we show that the DExD/H-box protein CYT-19, a general RNA chaperone, mediates ATP-dependent unfolding of both the native conformation and a long-lived misfolded conformation of a group I catalytic RNA with efficiencies that depend on the stabilities of the RNA species but not on specific structural features. CYT-19 then allows the RNA to refold, changing the distribution from equilibrium to kinetic control. Because misfolding is favoured kinetically, conditions that allow unfolding of the native RNA yield large increases in the population of misfolded species. Our results suggest that DExD/H-box proteins act with sufficient breadth and efficiency to allow structured RNAs to populate a wider range of conformations than would be present at equilibrium. Thus, RNAs may face selective pressure to stabilize their active conformations relative to inactive ones to avoid significant redistribution by DExD/H-box proteins. Conversely, RNAs whose functions depend on forming multiple conformations may rely on DExD/H-box proteins to increase the populations of less stable conformations, thereby increasing their overall efficiencies.
Figures
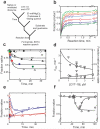
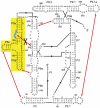
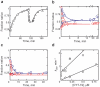
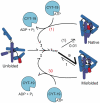
Comment in
-
Biochemistry: indifferent chaperones.Nature. 2007 Oct 25;449(7165):999-1000. doi: 10.1038/449999a. Nature. 2007. PMID: 17960232 No abstract available.
Similar articles
-
Nonspecific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone.Proc Natl Acad Sci U S A. 2006 Nov 7;103(45):16698-703. doi: 10.1073/pnas.0603127103. Epub 2006 Oct 30. Proc Natl Acad Sci U S A. 2006. PMID: 17075070 Free PMC article.
-
Biochemistry: indifferent chaperones.Nature. 2007 Oct 25;449(7165):999-1000. doi: 10.1038/449999a. Nature. 2007. PMID: 17960232 No abstract available.
-
Probing the mechanisms of DEAD-box proteins as general RNA chaperones: the C-terminal domain of CYT-19 mediates general recognition of RNA.Biochemistry. 2007 Mar 20;46(11):3013-22. doi: 10.1021/bi0619472. Epub 2007 Feb 21. Biochemistry. 2007. PMID: 17311413 Free PMC article.
-
Iterative annealing mechanism explains the functions of the GroEL and RNA chaperones.Protein Sci. 2020 Feb;29(2):360-377. doi: 10.1002/pro.3795. Epub 2019 Dec 23. Protein Sci. 2020. PMID: 31800116 Free PMC article. Review.
-
DEAD-box proteins as RNA helicases and chaperones.Wiley Interdiscip Rev RNA. 2011 Jan-Feb;2(1):135-52. doi: 10.1002/wrna.50. Wiley Interdiscip Rev RNA. 2011. PMID: 21297876 Free PMC article. Review.
Cited by
-
αβDCA method identifies unspecific binding but specific disruption of the group I intron by the StpA chaperone.RNA. 2020 Nov;26(11):1530-1540. doi: 10.1261/rna.074336.119. Epub 2020 Aug 3. RNA. 2020. PMID: 32747608 Free PMC article.
-
Modulating RNA structure and catalysis: lessons from small cleaving ribozymes.Cell Mol Life Sci. 2009 Dec;66(24):3937-50. doi: 10.1007/s00018-009-0124-1. Epub 2009 Aug 30. Cell Mol Life Sci. 2009. PMID: 19718544 Free PMC article. Review.
-
Solution structures of DEAD-box RNA chaperones reveal conformational changes and nucleic acid tethering by a basic tail.Proc Natl Acad Sci U S A. 2011 Jul 26;108(30):12254-9. doi: 10.1073/pnas.1109566108. Epub 2011 Jul 11. Proc Natl Acad Sci U S A. 2011. PMID: 21746911 Free PMC article.
-
Rapid RNA-ligand interaction analysis through high-information content conformational and stability landscapes.Nat Commun. 2015 Dec 7;6:8898. doi: 10.1038/ncomms9898. Nat Commun. 2015. PMID: 26638992 Free PMC article.
-
Mss116p: a DEAD-box protein facilitates RNA folding.RNA Biol. 2013 Jan;10(1):71-82. doi: 10.4161/rna.22492. Epub 2012 Oct 12. RNA Biol. 2013. PMID: 23064153 Free PMC article. Review.
References
References From Methods
-
- Herschlag D, Cech TR. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry. 1990;29:10159–10171. - PubMed
References
-
- Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell. 2001;8:251–262. - PubMed
-
- Jankowsky E, Fairman ME. RNA helicases - one fold for many functions. Curr. Opin. Struct. Biol. 2007;17:316–324. - PubMed
-
- Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 1993;3:419–429.
-
- Jankowsky E, Gross CH, Shuman S, Pyle AM. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

