Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone
- PMID: 17960235
- PMCID: PMC2581903
- DOI: 10.1038/nature06235
Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone
Abstract
DExD/H-box proteins are ubiquitously involved in RNA-mediated processes and use ATP to accelerate conformational changes in RNA. However, their mechanisms of action, and what determines which RNA species are targeted, are not well understood. Here we show that the DExD/H-box protein CYT-19, a general RNA chaperone, mediates ATP-dependent unfolding of both the native conformation and a long-lived misfolded conformation of a group I catalytic RNA with efficiencies that depend on the stabilities of the RNA species but not on specific structural features. CYT-19 then allows the RNA to refold, changing the distribution from equilibrium to kinetic control. Because misfolding is favoured kinetically, conditions that allow unfolding of the native RNA yield large increases in the population of misfolded species. Our results suggest that DExD/H-box proteins act with sufficient breadth and efficiency to allow structured RNAs to populate a wider range of conformations than would be present at equilibrium. Thus, RNAs may face selective pressure to stabilize their active conformations relative to inactive ones to avoid significant redistribution by DExD/H-box proteins. Conversely, RNAs whose functions depend on forming multiple conformations may rely on DExD/H-box proteins to increase the populations of less stable conformations, thereby increasing their overall efficiencies.
Figures
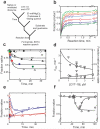
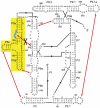
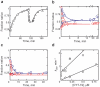
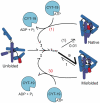
Comment in
-
Biochemistry: indifferent chaperones.Nature. 2007 Oct 25;449(7165):999-1000. doi: 10.1038/449999a. Nature. 2007. PMID: 17960232 No abstract available.
References
References From Methods
-
- Herschlag D, Cech TR. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry. 1990;29:10159–10171. - PubMed
References
-
- Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell. 2001;8:251–262. - PubMed
-
- Jankowsky E, Fairman ME. RNA helicases - one fold for many functions. Curr. Opin. Struct. Biol. 2007;17:316–324. - PubMed
-
- Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 1993;3:419–429.
-
- Jankowsky E, Gross CH, Shuman S, Pyle AM. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

