An elaborate pathway required for Ras-mediated epigenetic silencing
- PMID: 17960246
- PMCID: PMC2147719
- DOI: 10.1038/nature06251
An elaborate pathway required for Ras-mediated epigenetic silencing
Abstract
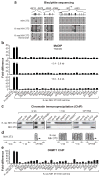
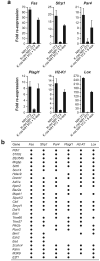
The conversion of a normal cell to a cancer cell occurs in several steps and typically involves the activation of oncogenes and the inactivation of tumour suppressor and pro-apoptotic genes. In many instances, inactivation of genes critical for cancer development occurs by epigenetic silencing, often involving hypermethylation of CpG-rich promoter regions. It remains to be determined whether silencing occurs by random acquisition of epigenetic marks that confer a selective growth advantage or through a specific pathway initiated by an oncogene. Here we perform a genome-wide RNA interference (RNAi) screen in K-ras-transformed NIH 3T3 cells and identify 28 genes required for Ras-mediated epigenetic silencing of the pro-apoptotic Fas gene. At least nine of these RESEs (Ras epigenetic silencing effectors), including the DNA methyltransferase DNMT1, are directly associated with specific regions of the Fas promoter in K-ras-transformed NIH 3T3 cells but not in untransformed NIH 3T3 cells. RNAi-mediated knockdown of any of the 28 RESEs results in failure to recruit DNMT1 to the Fas promoter, loss of Fas promoter hypermethylation, and derepression of Fas expression. Analysis of five other epigenetically repressed genes indicates that Ras directs the silencing of multiple unrelated genes through a largely common pathway. Last, we show that nine RESEs are required for anchorage-independent growth and tumorigenicity of K-ras-transformed NIH 3T3 cells; these nine genes have not previously been implicated in transformation by Ras. Our results show that Ras-mediated epigenetic silencing occurs through a specific, complex, pathway involving components that are required for maintenance of a fully transformed phenotype.
Figures
Similar articles
-
Oncogenic RAS directs silencing of tumor suppressor genes through ordered recruitment of transcriptional repressors.Genes Dev. 2013 Oct 15;27(20):2221-6. doi: 10.1101/gad.227413.113. Epub 2013 Oct 8. Genes Dev. 2013. PMID: 24105743 Free PMC article.
-
RAS-mediated epigenetic inactivation of OPCML in oncogenic transformation of human ovarian surface epithelial cells.FASEB J. 2006 Mar;20(3):497-9. doi: 10.1096/fj.05-4586fje. Epub 2005 Dec 29. FASEB J. 2006. PMID: 16384911
-
PEA3 sites within the progression elevated gene-3 (PEG-3) promoter and mitogen-activated protein kinase contribute to differential PEG-3 expression in Ha-ras and v-raf oncogene transformed rat embryo cells.Nucleic Acids Res. 2001 Apr 15;29(8):1661-71. doi: 10.1093/nar/29.8.1661. Nucleic Acids Res. 2001. PMID: 11292838 Free PMC article.
-
Gene methylation in gastric cancer.Clin Chim Acta. 2013 Sep 23;424:53-65. doi: 10.1016/j.cca.2013.05.002. Epub 2013 May 10. Clin Chim Acta. 2013. PMID: 23669186 Review.
-
The role of non-ras transforming genes in chemical carcinogenesis.Environ Health Perspect. 1991 Jun;93:33-40. doi: 10.1289/ehp.919333. Environ Health Perspect. 1991. PMID: 1685444 Free PMC article. Review.
Cited by
-
Resistance to therapy in BRCA2 mutant cells due to loss of the nucleosome remodeling factor CHD4.Genes Dev. 2015 Mar 1;29(5):489-94. doi: 10.1101/gad.256214.114. Genes Dev. 2015. PMID: 25737278 Free PMC article.
-
HOXC6 drives a therapeutically targetable pancreatic cancer growth and metastasis pathway by regulating MSK1 and PPP2R2B.Cell Rep Med. 2023 Nov 21;4(11):101285. doi: 10.1016/j.xcrm.2023.101285. Epub 2023 Nov 10. Cell Rep Med. 2023. PMID: 37951219 Free PMC article.
-
Loss of ARF sensitizes transgenic BRAFV600E mice to UV-induced melanoma via suppression of XPC.Cancer Res. 2013 Jul 15;73(14):4337-48. doi: 10.1158/0008-5472.CAN-12-4454. Epub 2013 May 6. Cancer Res. 2013. PMID: 23650282 Free PMC article.
-
The histone H3K9 demethylase KDM3A promotes anoikis by transcriptionally activating pro-apoptotic genes BNIP3 and BNIP3L.Elife. 2016 Jul 29;5:e16844. doi: 10.7554/eLife.16844. Elife. 2016. PMID: 27472901 Free PMC article.
-
Two Dot1 isoforms in Saccharomyces cerevisiae as a result of leaky scanning by the ribosome.Nucleic Acids Res. 2009 Nov;37(21):7047-58. doi: 10.1093/nar/gkp765. Nucleic Acids Res. 2009. PMID: 19778927 Free PMC article.
References
-
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. - PubMed
-
- Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2(Suppl 1):S4–11. - PubMed
-
- Jones PA. DNA methylation errors and cancer. Cancer Res. 1996;56:2463–2467. - PubMed
-
- Baylin S, Bestor TH. Altered methylation patterns in cancer cell genomes: cause or consequence? Cancer Cell. 2002;1:299–305. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

