Mutational analysis of the purine riboswitch aptamer domain
- PMID: 17960911
- PMCID: PMC2556308
- DOI: 10.1021/bi700410g
Mutational analysis of the purine riboswitch aptamer domain
Abstract
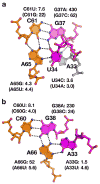
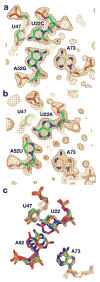
The purine riboswitch is one of a number of mRNA elements commonly found in the 5'-untranslated region capable of controlling expression in a cis-fashion via its ability to directly bind small-molecule metabolites. Extensive biochemical and structural analysis of the nucleobase-binding domain of the riboswitch, referred to as the aptamer domain, has revealed that the mRNA recognizes its cognate ligand using an intricately folded three-way junction motif that completely encapsulates the ligand. High-affinity binding of the purine nucleobase is facilitated by a distal loop-loop interaction that is conserved between both the adenine and guanine riboswitches. To understand the contribution of conserved nucleotides in both the three-way junction and the loop-loop interaction of this RNA, we performed a detailed mutagenic survey of these elements in the context of an adenine-responsive variant of the xpt-pbuX guanine riboswitch from Bacillus subtilis. The varying ability of these mutants to bind ligand as measured by isothermal titration calorimetry uncovered the conserved nucleotides whose identity is required for purine binding. Crystallographic analysis of the bound form of five mutants and chemical probing of their free state demonstrate that the identity of several universally conserved nucleotides is not essential for formation of the RNA-ligand complex but rather for maintaining a binding-competent form of the free RNA. These data show that conservation patterns in riboswitches arise from a combination of formation of the ligand-bound complex, promoting an open form of the free RNA, and participating in the secondary structural switch with the expression platform.
Figures
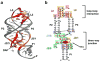




Similar articles
-
Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain.J Mol Biol. 2006 Jun 9;359(3):754-68. doi: 10.1016/j.jmb.2006.04.003. Epub 2006 Apr 21. J Mol Biol. 2006. PMID: 16650860
-
Interplay of 'induced fit' and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch.Nucleic Acids Res. 2007;35(2):572-83. doi: 10.1093/nar/gkl1094. Epub 2006 Dec 14. Nucleic Acids Res. 2007. PMID: 17175531 Free PMC article.
-
Ligand-dependent folding of the three-way junction in the purine riboswitch.RNA. 2008 Apr;14(4):675-84. doi: 10.1261/rna.736908. Epub 2008 Feb 11. RNA. 2008. PMID: 18268025 Free PMC article.
-
Structural studies of the purine and SAM binding riboswitches.Cold Spring Harb Symp Quant Biol. 2006;71:259-68. doi: 10.1101/sqb.2006.71.015. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381305 Review.
-
Purine sensing by riboswitches.Biol Cell. 2008 Jan;100(1):1-11. doi: 10.1042/BC20070088. Biol Cell. 2008. PMID: 18072940 Review.
Cited by
-
'Z-DNA like' fragments in RNA: a recurring structural motif with implications for folding, RNA/protein recognition and immune response.Nucleic Acids Res. 2016 Jul 8;44(12):5944-56. doi: 10.1093/nar/gkw388. Epub 2016 May 5. Nucleic Acids Res. 2016. PMID: 27151194 Free PMC article.
-
Signal amplification and optimization of riboswitch-based hybrid inputs by modular and titratable toehold switches.J Biol Eng. 2021 Mar 19;15(1):11. doi: 10.1186/s13036-021-00261-w. J Biol Eng. 2021. PMID: 33741029 Free PMC article.
-
Multivector fluorescence analysis of the xpt guanine riboswitch aptamer domain and the conformational role of guanine.Biochemistry. 2010 Mar 2;49(8):1596-605. doi: 10.1021/bi9019912. Biochemistry. 2010. PMID: 20108980 Free PMC article.
-
Molecular insights into the ligand-controlled organization of the SAM-I riboswitch.Nat Chem Biol. 2011 Jun;7(6):384-92. doi: 10.1038/nchembio.563. Epub 2011 May 1. Nat Chem Biol. 2011. PMID: 21532599
-
Constitutive regulatory activity of an evolutionarily excluded riboswitch variant.J Biol Chem. 2011 Aug 5;286(31):27406-15. doi: 10.1074/jbc.M111.229047. Epub 2011 Jun 15. J Biol Chem. 2011. PMID: 21676871 Free PMC article.
References
-
- Costa FF. Non-coding RNAs: Lost in translation? Gene. 2007;386:1–10. - PubMed
-
- Storz G. An expanding universe of noncoding RNAs. Science. 2002;296:1260–3. - PubMed
-
- Storz G, Altuvia S, Wassarman KM. An abundance of RNA regulators. Annu Rev Biochem. 2005;74:199–217. - PubMed
-
- Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. A new frontier for molecular medicine: noncoding RNAs. Biochim Biophys Acta. 2005;1756:65–75. - PubMed
-
- Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Curr Opin Struct Biol. 2005;15:342–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

