Dicyema Pax6 and Zic: tool-kit genes in a highly simplified bilaterian
- PMID: 17961212
- PMCID: PMC2222250
- DOI: 10.1186/1471-2148-7-201
Dicyema Pax6 and Zic: tool-kit genes in a highly simplified bilaterian
Abstract
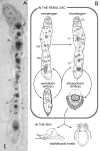
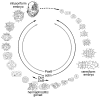
Background: Dicyemid mesozoans (Phylum Dicyemida) are simple (8-40-cell) cephalopod endoparasites. They have neither body cavities nor differentiated organs, such as nervous and gastrointestinal systems. Whether dicyemids are intermediate between Protozoa and Metazoa (as represented by their "Mesozoa" classification) or degenerate species of more complex metazoans is controversial. Recent molecular phylogenetic studies suggested that they are simplified bilaterians belonging to the Lophotrochozoa. We cloned two genes developmentally critical in bilaterian animals (Pax6 and Zic), together with housekeeping genes (actin, fructose-bisphosphate aldolase, and ATP synthase beta subunit) from a dicyemid to reveal whether their molecular phylogeny supported the "simplification" hypothesis, and to clarify evolutionary changes in dicyemid gene structure and expression profiles.
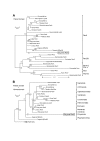
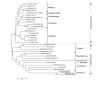
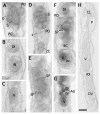
Results: Genomic/cDNA sequence analysis showed that 1) the Pax6 molecular phylogeny and Zic intron positions supported the idea of dicyemids as reduced bilaterians; 2) the aa sequences deduced from the five genes were highly divergent; and 3) Dicyema genes contained very short introns of uniform length. In situ hybridization analyses revealed that Zic genes were expressed in hermaphroditic gonads, and Pax6 was expressed weakly throughout the developmental stages of the 2 types of embryo and in the hermaphroditic gonads.
Conclusion: The accelerated evolutionary rates and very short and uniform intron may represent a part of Dicyema genomic features. The presence and expression of the two tool-kit genes (Pax6 and Zic) in Dicyema suggests that they can be very versatile genes even required for the highly reduced bilaterian like Dicyema. Dicyemids may be useful models of evolutionary body plan simplification.
Figures



Similar articles
-
The expression of tubulin and tektin genes in dicyemid mesozoans (Phylum: Dicyemida).J Parasitol. 2007 Jun;93(3):608-18. doi: 10.1645/GE-1037R.1. J Parasitol. 2007. PMID: 17626353
-
Cloning of chitinase-like protein1 cDNA from dicyemid mesozoans (Phylum: Dicyemida).J Parasitol. 2007 Dec;93(6):1403-15. doi: 10.1645/GE-1290.1. J Parasitol. 2007. PMID: 18314687
-
Phylogenetic position of the dicyemid mesozoa inferred from 18S rDNA sequences.Biol Bull. 1995 Oct-Nov;189(2):81-90. doi: 10.2307/1542458. Biol Bull. 1995. PMID: 8541419
-
Lophotrochozoan Zic Genes.Adv Exp Med Biol. 2018;1046:69-86. doi: 10.1007/978-981-10-7311-3_5. Adv Exp Med Biol. 2018. PMID: 29442318 Review.
-
The evolution of vision.Wiley Interdiscip Rev Dev Biol. 2014 Jan-Feb;3(1):1-40. doi: 10.1002/wdev.96. Epub 2012 Dec 21. Wiley Interdiscip Rev Dev Biol. 2014. PMID: 24902832 Review.
Cited by
-
Role of BMP, FGF, calcium signaling, and Zic proteins in vertebrate neuroectodermal differentiation.Neurochem Res. 2011 Jul;36(7):1286-92. doi: 10.1007/s11064-011-0422-5. Epub 2011 Feb 19. Neurochem Res. 2011. PMID: 21336820 Free PMC article. Review.
-
Expression pattern of annelid Zic in embryonic development of the oligochaete Tubifex tubifex.Dev Genes Evol. 2008 Oct;218(10):553-60. doi: 10.1007/s00427-008-0252-x. Epub 2008 Sep 23. Dev Genes Evol. 2008. PMID: 18810489
-
Five new species of dicyemid mesozoans (Dicyemida: Dicyemidae) from two Australian cuttlefish species, with comments on dicyemid fauna composition.Syst Parasitol. 2013 Oct;86(2):125-51. doi: 10.1007/s11230-013-9443-6. Epub 2013 Sep 19. Syst Parasitol. 2013. PMID: 24048746
-
Dicyemida and Orthonectida: Two Stories of Body Plan Simplification.Front Genet. 2019 May 24;10:443. doi: 10.3389/fgene.2019.00443. eCollection 2019. Front Genet. 2019. PMID: 31178892 Free PMC article.
-
Constrained intron structures in a microsporidian.Mol Biol Evol. 2010 Sep;27(9):1979-82. doi: 10.1093/molbev/msq087. Epub 2010 Apr 1. Mol Biol Evol. 2010. PMID: 20360213 Free PMC article.
References
-
- Gehring WJ. The genetic control of eye development and its implications for the evolution of the various eye-types. Int J Dev Biol. 2002;46:65–73. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

