Regulation of poly(ADP-ribose) polymerase-1 (PARP-1) gene expression through the post-translational modification of Sp1: a nuclear target protein of PARP-1
- PMID: 17961220
- PMCID: PMC2175517
- DOI: 10.1186/1471-2199-8-96
Regulation of poly(ADP-ribose) polymerase-1 (PARP-1) gene expression through the post-translational modification of Sp1: a nuclear target protein of PARP-1
Abstract
Background: Poly(ADP-ribose) polymerase-1 (PARP-1) is a nuclear enzyme that plays critical functions in many biological processes, including DNA repair and gene transcription. The main function of PARP-1 is to catalyze the transfer of ADP-ribose units from nicotinamide adenine dinucleotide (NAD+) to a large array of acceptor proteins, which comprises histones, transcription factors, as well as PARP-1 itself. We have previously demonstrated that transcription of the PARP-1 gene essentially rely on the opposite regulatory actions of two distinct transcription factors, Sp1 and NFI. In the present study, we examined whether suppression of PARP-1 expression in embryonic fibroblasts derived from PARP-1 knockout mice (PARP-1-/-) might alter the expression and/or DNA binding properties of Sp1 and NFI. We also explored the possibility that Sp1 or NFI (or both) may represent target proteins of PARP-1 activity.
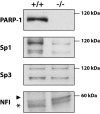
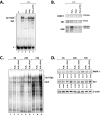
Results: Expression of both Sp1 and NFI was found to be considerably reduced in PARP-1-/- cells. Co-immunoprecipitation assays revealed that PARP-1 physically interacts with Sp1 in a DNA-independent manner, but neither with Sp3 nor NFI, in PARP-1+/+ cells. In addition, in vitro PARP assays indicated that PARP-1 could catalyze the addition of polymer of ADP-ribose to Sp1, which also translated into a reduction of Sp1 binding to its consensus DNA target site. Transfection of the PARP-1 promoter into both PARP-1+/+ and PARP-1-/- cells revealed that the lack of PARP-1 expression in PARP-1-/- cells also results in a strong increase in PARP-1 promoter activity. This influence of PARP-1 was found to rely on the presence of the Sp1 sites present on the basal PARP-1 promoter as their mutation entirely abolished the increased promoter activity observed in PARP-1-/- cells. Subjecting PARP-1+/+ cells to an oxidative challenge with hydrogen peroxide to increase PARP-1 activity translated into a dramatic reduction in the DNA binding properties of Sp1. However, its suppression by the inhibitor PJ34 improved DNA binding of Sp1 and led to a dramatic increase in PARP-1 promoter function.
Conclusion: Our results therefore recognized Sp1 as a target protein of PARP-1 activity, the addition of polymer of ADP-ribose to this transcription factor restricting its positive regulatory influence on gene transcription.
Figures







Similar articles
-
Expression of the gene encoding poly(ADP-ribose) polymerase-1 is modulated by fibronectin during corneal wound healing.Invest Ophthalmol Vis Sci. 2006 Oct;47(10):4199-210. doi: 10.1167/iovs.06-0176. Invest Ophthalmol Vis Sci. 2006. PMID: 17003407
-
Regulation of the poly(ADP-ribose) polymerase-1 gene expression by the transcription factors Sp1 and Sp3 is under the influence of cell density in primary cultured cells.Biochem J. 2005 Jul 15;389(Pt 2):423-33. doi: 10.1042/BJ20041718. Biochem J. 2005. PMID: 15777284 Free PMC article.
-
A nuclear factor other than Sp1 binds the GC-rich promoter of the gene encoding rat poly(ADP-ribose) polymerase in vitro.Biochem Cell Biol. 1997;75(4):427-34. Biochem Cell Biol. 1997. PMID: 9493965
-
Poly(ADP-ribose): PARadigms and PARadoxes.Mol Aspects Med. 2013 Dec;34(6):1046-65. doi: 10.1016/j.mam.2012.12.010. Epub 2013 Jan 2. Mol Aspects Med. 2013. PMID: 23290998 Review.
-
Structure and function of poly(ADP-ribose) polymerase-1: role in oxidative stress-related pathologies.Curr Vasc Pharmacol. 2005 Jul;3(3):209-14. doi: 10.2174/1570161054368625. Curr Vasc Pharmacol. 2005. PMID: 16026317 Review.
Cited by
-
Contribution of the Transcription Factors Sp1/Sp3 and AP-1 to Clusterin Gene Expression during Corneal Wound Healing of Tissue-Engineered Human Corneas.Int J Mol Sci. 2021 Nov 17;22(22):12426. doi: 10.3390/ijms222212426. Int J Mol Sci. 2021. PMID: 34830308 Free PMC article.
-
PARP1 poly(ADP-ribosyl)ates Sox2 to control Sox2 protein levels and FGF4 expression during embryonic stem cell differentiation.J Biol Chem. 2009 Aug 14;284(33):22263-22273. doi: 10.1074/jbc.M109.033118. Epub 2009 Jun 16. J Biol Chem. 2009. PMID: 19531481 Free PMC article.
-
Poly(ADP-ribose) Polyremase-1 (PARP-1) Inhibition: A Promising Therapeutic Strategy for ETS-Expressing Tumours.Int J Mol Sci. 2023 Aug 30;24(17):13454. doi: 10.3390/ijms241713454. Int J Mol Sci. 2023. PMID: 37686260 Free PMC article. Review.
-
Positive transcriptional regulation of the human micro opioid receptor gene by poly(ADP-ribose) polymerase-1 and increase of its DNA binding affinity based on polymorphism of G-172 -> T.J Biol Chem. 2009 Jul 24;284(30):20175-83. doi: 10.1074/jbc.M109.019414. Epub 2009 May 15. J Biol Chem. 2009. PMID: 19447888 Free PMC article.
-
Mechanical stretch induces angiotensinogen expression through PARP1 activation in kidney proximal tubular cells.In Vitro Cell Dev Biol Anim. 2015 Jan;51(1):72-8. doi: 10.1007/s11626-014-9809-3. Epub 2014 Aug 23. In Vitro Cell Dev Biol Anim. 2015. PMID: 25148826
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

