Structure of Arabidopsis thaliana 5-methylthioribose kinase reveals a more occluded active site than its bacterial homolog
- PMID: 17961230
- PMCID: PMC2194712
- DOI: 10.1186/1472-6807-7-70
Structure of Arabidopsis thaliana 5-methylthioribose kinase reveals a more occluded active site than its bacterial homolog
Abstract
Background: Metabolic variations exist between the methionine salvage pathway of humans and a number of plants and microbial pathogens. 5-Methylthioribose (MTR) kinase is a key enzyme required for methionine salvage in plants and many bacteria. The absence of a mammalian homolog suggests that MTR kinase is a good target for the design of specific herbicides or antibiotics.
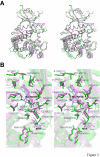
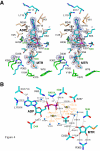
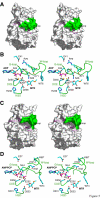
Results: The structure of Arabidopsis thaliana MTR kinase co-crystallized with ATPgammaS and MTR has been determined at 1.9 A resolution. The structure is similar to B. subtilis MTR kinase and has the same protein kinase fold observed in other evolutionarily related protein kinase-like phosphotransferases. The active site is comparable between the two enzymes with the DXE-motif coordinating the nucleotide-Mg, the D238 of the HGD catalytic loop polarizing the MTR O1 oxygen, and the RR-motif interacting with the substrate MTR. Unlike its bacterial homolog, however, the Gly-rich loop (G-loop) of A. thaliana MTR kinase has an extended conformation, which shields most of the active site from solvent, a feature that resembles eukaryotic protein kinases more than the bacterial enzyme. The G- and W-loops of A. thaliana and B. subtilis MTR kinase adopt different conformations despite high sequence similarity. The ATPgammaS analog was hydrolyzed during the co-crystallization procedure, resulting in ADP in the active site. This suggests that the A. thaliana enzyme, like its bacterial homolog, may have significant ATPase activity in the absence of MTR.
Conclusion: The structure of A. thaliana MTR kinase provides a template for structure-based design of agrochemicals, particularly herbicides whose effectiveness could be regulated by nutrient levels. Features of the MTR binding site offer an opportunity for a simple organic salt of an MTR analog to specifically inhibit MTR kinase.
Figures


Similar articles
-
Structures of 5-methylthioribose kinase reveal substrate specificity and unusual mode of nucleotide binding.J Biol Chem. 2007 Jul 27;282(30):22195-206. doi: 10.1074/jbc.M611045200. Epub 2007 May 23. J Biol Chem. 2007. PMID: 17522047
-
Crystallization and preliminary X-ray analysis of 5'-methylthioribose kinase from Bacillus subtilis and Arabidopsis thaliana.Acta Crystallogr D Biol Crystallogr. 2004 Jan;60(Pt 1):116-9. doi: 10.1107/s0907444903022042. Epub 2003 Dec 18. Acta Crystallogr D Biol Crystallogr. 2004. PMID: 14684902
-
ADP-2Ho as a phasing tool for nucleotide-containing proteins.Acta Crystallogr D Biol Crystallogr. 2007 Apr;63(Pt 4):493-9. doi: 10.1107/S0907444907006592. Epub 2007 Mar 16. Acta Crystallogr D Biol Crystallogr. 2007. PMID: 17372354
-
Structure and function of cellular deoxyribonucleoside kinases.Cell Mol Life Sci. 2002 Aug;59(8):1327-46. doi: 10.1007/s00018-002-8511-x. Cell Mol Life Sci. 2002. PMID: 12363036 Free PMC article. Review.
-
Small substrate, big surprise: fold, function and phylogeny of dihydroxyacetone kinases.Cell Mol Life Sci. 2006 Apr;63(7-8):890-900. doi: 10.1007/s00018-005-5518-0. Cell Mol Life Sci. 2006. PMID: 16505971 Free PMC article. Review.
Cited by
-
Phloem-specific expression of Yang cycle genes and identification of novel Yang cycle enzymes in Plantago and Arabidopsis.Plant Cell. 2011 May;23(5):1904-19. doi: 10.1105/tpc.110.079657. Epub 2011 May 3. Plant Cell. 2011. PMID: 21540433 Free PMC article.
-
Identification and classification of small molecule kinases: insights into substrate recognition and specificity.BMC Evol Biol. 2016 Jan 6;16:7. doi: 10.1186/s12862-015-0576-x. BMC Evol Biol. 2016. PMID: 26738562 Free PMC article.
-
Substrate recognition by the 4-hydroxytryptamine kinase PsiK in psilocybin biosynthesis.FEBS Lett. 2025 Feb;599(3):447-455. doi: 10.1002/1873-3468.15042. Epub 2024 Oct 24. FEBS Lett. 2025. PMID: 39449146 Free PMC article.
-
Structural basis for psilocybin biosynthesis.Nat Commun. 2025 Mar 22;16(1):2827. doi: 10.1038/s41467-025-58239-x. Nat Commun. 2025. PMID: 40121242 Free PMC article.
-
Structure of the antibiotic resistance factor spectinomycin phosphotransferase from Legionella pneumophila.J Biol Chem. 2010 Mar 26;285(13):9545-9555. doi: 10.1074/jbc.M109.038364. Epub 2010 Jan 19. J Biol Chem. 2010. PMID: 20089863 Free PMC article.
References
-
- Roje S. S-Adenosyl-l-methionine: Beyond the universal methyl group donor. Phytochemistry. 2006 - PubMed
-
- Clarke S, Banfield K. S-adenosylmethionine-dependent methyltransferase. In: Carmel R, Jacobsen DW, editor. Homocysteine in Health and Disease. New York , Cambridge University Press; 2001. pp. 63–78.
-
- Pegg AE. Polyamine metabolism and its importance in neoplastic growth and a target for chemotherapy. Cancer Res. 1988;48:759–774. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

