Whole genome linkage disequilibrium maps in cattle
- PMID: 17961247
- PMCID: PMC2174945
- DOI: 10.1186/1471-2156-8-74
Whole genome linkage disequilibrium maps in cattle
Abstract
Background: Bovine whole genome linkage disequilibrium maps were constructed for eight breeds of cattle. These data provide fundamental information concerning bovine genome organization which will allow the design of studies to associate genetic variation with economically important traits and also provides background information concerning the extent of long range linkage disequilibrium in cattle.
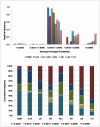
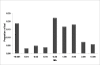
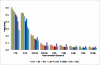
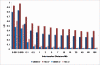
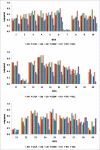
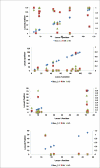
Results: Linkage disequilibrium was assessed using r2 among all pairs of syntenic markers within eight breeds of cattle from the Bos taurus and Bos indicus subspecies. Bos taurus breeds included Angus, Charolais, Dutch Black and White Dairy, Holstein, Japanese Black and Limousin while Bos indicus breeds included Brahman and Nelore. Approximately 2670 markers spanning the entire bovine autosomal genome were used to estimate pairwise r2 values. We found that the extent of linkage disequilibrium is no more than 0.5 Mb in these eight breeds of cattle.
Conclusion: Linkage disequilibrium in cattle has previously been reported to extend several tens of centimorgans. Our results, based on a much larger sample of marker loci and across eight breeds of cattle indicate that in cattle linkage disequilibrium persists over much more limited distances. Our findings suggest that 30,000-50,000 loci will be needed to conduct whole genome association studies in cattle.
Figures

References
-
- Vallejo RL, Li YL, Rogers GW, Ashwell MS. Genetic diversity and background linkage disequilibrium in the North American Holstein cattle population. J Dairy Sci. 2003;86:4137–4147. - PubMed
-
- Tenesa A, Knott SA, Ward D, Smith D, Williams JL, Visscher PM. Estimation of linkage disequilibrium in a sample of the United Kingdom dairy cattle population using unphased genotypes. J Anim Sci. 2003;81:617–623. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

