The challenge of protein structure determination--lessons from structural genomics
- PMID: 17962404
- PMCID: PMC2211687
- DOI: 10.1110/ps.073037907
The challenge of protein structure determination--lessons from structural genomics
Abstract
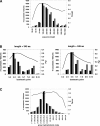
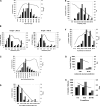
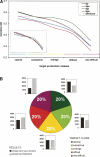
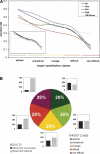
The process of experimental determination of protein structure is marred with a high ratio of failures at many stages. With availability of large quantities of data from high-throughput structure determination in structural genomics centers, we can now learn to recognize protein features correlated with failures; thus, we can recognize proteins more likely to succeed and eventually learn how to modify those that are less likely to succeed. Here, we identify several protein features that correlate strongly with successful protein production and crystallization and combine them into a single score that assesses "crystallization feasibility." The formula derived here was tested with a jackknife procedure and validated on independent benchmark sets. The "crystallization feasibility" score described here is being applied to target selection in the Joint Center for Structural Genomics, and is now contributing to increasing the success rate, lowering the costs, and shortening the time for protein structure determination. Analyses of PDB depositions suggest that very similar features also play a role in non-high-throughput structure determination, suggesting that this crystallization feasibility score would also be of significant interest to structural biology, as well as to molecular and biochemistry laboratories.
Figures

Similar articles
-
The impact of structural genomics: the first quindecennial.J Struct Funct Genomics. 2016 Mar;17(1):1-16. doi: 10.1007/s10969-016-9201-5. Epub 2016 Mar 2. J Struct Funct Genomics. 2016. PMID: 26935210 Free PMC article. Review.
-
NMR in structural genomics to increase structural coverage of the protein universe: Delivered by Prof. Kurt Wüthrich on 7 July 2013 at the 38th FEBS Congress in St. Petersburg, Russia.FEBS J. 2016 Nov;283(21):3870-3881. doi: 10.1111/febs.13751. Epub 2016 Jun 9. FEBS J. 2016. PMID: 27154589 Free PMC article.
-
A tour of structural genomics.Nat Rev Genet. 2001 Oct;2(10):801-9. doi: 10.1038/35093574. Nat Rev Genet. 2001. PMID: 11584296 Review.
-
Protein Crystallizability.Methods Mol Biol. 2016;1415:341-70. doi: 10.1007/978-1-4939-3572-7_17. Methods Mol Biol. 2016. PMID: 27115641 Review.
-
Strategies for improving crystallization success rates.Methods Mol Biol. 2008;426:345-62. doi: 10.1007/978-1-60327-058-8_22. Methods Mol Biol. 2008. PMID: 18542875
Cited by
-
Sequence-based prediction of protein crystallization, purification and production propensity.Bioinformatics. 2011 Jul 1;27(13):i24-33. doi: 10.1093/bioinformatics/btr229. Bioinformatics. 2011. PMID: 21685077 Free PMC article.
-
Functional consequences of somatic mutations in cancer using protein pocket-based prioritization approach.Genome Med. 2014 Oct 14;6(10):81. doi: 10.1186/s13073-014-0081-7. eCollection 2014. Genome Med. 2014. PMID: 25360158 Free PMC article.
-
ImmuneBuilder: Deep-Learning models for predicting the structures of immune proteins.Commun Biol. 2023 May 29;6(1):575. doi: 10.1038/s42003-023-04927-7. Commun Biol. 2023. PMID: 37248282 Free PMC article.
-
Is unphosphorylated Rex, as multifunctional protein of HTLV-1, a fully intrinsically disordered protein? An in silico study.Biochem Biophys Rep. 2016 Aug 4;8:14-22. doi: 10.1016/j.bbrep.2016.07.018. eCollection 2016 Dec. Biochem Biophys Rep. 2016. PMID: 28955936 Free PMC article.
-
Combining Wet and Dry Lab Techniques to Guide the Crystallization of Large Coiled-coil Containing Proteins.J Vis Exp. 2017 Jan 6;(119):54886. doi: 10.3791/54886. J Vis Exp. 2017. PMID: 28117766 Free PMC article.
References
-
- Bendtsen J.D., Nielsen, H., von Heijne, G., and Brunak, S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340: 783–795. - PubMed
-
- Bertone P., Kluger, Y., Lan, N., Zheng, D., Christendat, D., Yee, A., Edwards, A.M., Arrowsmith, C.H., Montelione, G.T., and Gerstein, M. 2001. SPINE: An integrated tracking database and data mining approach for identifying feasible targets in high-throughput structural proteomics. Nucleic Acids Res. 29: 2884–2898. - PMC - PubMed
-
- Canaves J.M., Page, R., Wilson, I.A., and Stevens, R.C. 2004. Protein biophysical properties that correlate with crystallization success in Thermotoga maritima: Maximum clustering strategy for structural genomics. J. Mol. Biol. 344: 977–991. - PubMed
-
- Chandonia J.M., Kim, S.H., and Brenner, S.E. 2006. Target selection and deselection at the Berkeley Structural Genomics Center. Proteins 62: 356–370. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

