Influence of oligomerization on the dynamics of G-protein coupled receptors as assessed by normal mode analysis
- PMID: 17963239
- PMCID: PMC3489929
- DOI: 10.1002/prot.21787
Influence of oligomerization on the dynamics of G-protein coupled receptors as assessed by normal mode analysis
Abstract
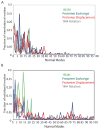
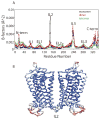
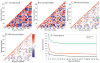
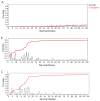
The recently discovered impact of oligomerization on G-protein coupled receptor (GPCR) function further complicates the already challenging goal of unraveling the molecular and dynamic mechanisms of these receptors. To help understand the effect of oligomerization on the dynamics of GPCRs, we have compared the motion of monomeric, dimeric, and tetrameric arrangements of the prototypic GPCR rhodopsin, using an approximate-yet powerful-normal mode analysis (NMA) technique termed elastic network model (ENM). Moreover, we have used ENM to discriminate between putative dynamic mechanisms likely to account for the recently observed conformational rearrangement of the TM4,5-TM4,5 dimerization interface of GPCRs that occurs upon activation. Our results indicate: (1) significant perturbation of the normal modes (NMs) of the rhodopsin monomer upon oligomerization, which is mainly manifested at interfacial regions; (2) increased positive correlation among the transmembrane domains (TMs) and between the extracellular loop (EL) and TM regions of the rhodopsin protomer; (3) highest interresidue positive correlation at the interfaces between protomers; and (4) experimentally testable hypotheses of differential motional changes within different putative oligomeric arrangements.
Figures



References
-
- Muller G. Towards 3D structures of G protein-coupled receptors: a multidisciplinary approach. Curr Med Chem. 2000;7:861–888. - PubMed
-
- George SR, O’Dowd BF, Lee SP. G-protein-coupled receptor oligomerization and its potential for drug discovery. Nat Rev Drug Discov. 2002;1:808–820. - PubMed
-
- Filizola M, Weinstein H. The structure and dynamics of GPCR oligomers: a new focus in models of cell-signaling mechanisms and drug design. Curr Opin Drug Discov Dev. 2005;8:577–584. - PubMed
-
- Milligan G. G-protein-coupled receptor heterodimers: pharmacology, function and relevance to drug discovery. Drug Discov Today. 2006;11:541–549. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

