Repeat-protein folding: new insights into origins of cooperativity, stability, and topology
- PMID: 17963718
- PMCID: PMC2474553
- DOI: 10.1016/j.abb.2007.08.034
Repeat-protein folding: new insights into origins of cooperativity, stability, and topology
Abstract
Although our understanding of globular protein folding continues to advance, the irregular tertiary structures and high cooperativity of globular proteins complicates energetic dissection. Recently, proteins with regular, repetitive tertiary structures have been identified that sidestep limitations imposed by globular protein architecture. Here we review recent studies of repeat-protein folding. These studies uniquely advance our understanding of both the energetics and kinetics of protein folding. Equilibrium studies provide detailed maps of local stabilities, access to energy landscapes, insights into cooperativity, determination of nearest-neighbor interaction parameters using statistical thermodynamics, relationships between consensus sequences and repeat-protein stability. Kinetic studies provide insight into the influence of short-range topology on folding rates, the degree to which folding proceeds by parallel (versus localized) pathways, and the factors that select among multiple potential pathways. The recent application of force spectroscopy to repeat-protein unfolding is providing a unique route to test and extend many of these findings.
Figures

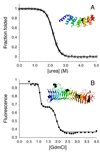

Similar articles
-
Cooperativity, smooth energy landscapes and the origins of topology-dependent protein folding rates.J Mol Biol. 2003 Feb 7;326(1):247-53. doi: 10.1016/s0022-2836(02)01356-6. J Mol Biol. 2003. PMID: 12547206
-
Analysis of Tandem Repeat Protein Folding Using Nearest-Neighbor Models.Annu Rev Biophys. 2021 May 6;50:245-265. doi: 10.1146/annurev-biophys-102220-083020. Epub 2021 Feb 19. Annu Rev Biophys. 2021. PMID: 33606943 Free PMC article. Review.
-
Dissecting and reprogramming the folding and assembly of tandem-repeat proteins.Biochem Soc Trans. 2015 Oct;43(5):881-8. doi: 10.1042/BST20150099. Biochem Soc Trans. 2015. PMID: 26517898 Review.
-
Modulation of folding kinetics of repeat proteins: interplay between intra- and interdomain interactions.Biophys J. 2012 Oct 3;103(7):1555-65. doi: 10.1016/j.bpj.2012.08.018. Epub 2012 Oct 2. Biophys J. 2012. PMID: 23062348 Free PMC article.
-
Exploring the folding energy landscape of a series of designed consensus tetratricopeptide repeat proteins.Proc Natl Acad Sci U S A. 2009 Oct 13;106(41):17383-8. doi: 10.1073/pnas.0907455106. Epub 2009 Oct 1. Proc Natl Acad Sci U S A. 2009. PMID: 19805120 Free PMC article.
Cited by
-
Modulating repeat protein stability: the effect of individual helix stability on the collective behavior of the ensemble.Protein Sci. 2011 Jun;20(6):1042-7. doi: 10.1002/pro.638. Epub 2011 May 3. Protein Sci. 2011. PMID: 21495096 Free PMC article.
-
A Naturally Occurring Repeat Protein with High Internal Sequence Identity Defines a New Class of TPR-like Proteins.Structure. 2015 Nov 3;23(11):2055-65. doi: 10.1016/j.str.2015.07.022. Epub 2015 Oct 1. Structure. 2015. PMID: 26439765 Free PMC article.
-
Artificially designed recombinant protein composed of multiple epitopes of foot-and-mouth disease virus as a vaccine candidate.Microb Cell Fact. 2017 Feb 22;16(1):33. doi: 10.1186/s12934-017-0648-2. Microb Cell Fact. 2017. PMID: 28228147 Free PMC article.
-
Direct observation of parallel folding pathways revealed using a symmetric repeat protein system.Biophys J. 2014 Jul 1;107(1):220-32. doi: 10.1016/j.bpj.2014.04.058. Biophys J. 2014. PMID: 24988356 Free PMC article.
-
The leucine-rich repeat domain of Internalin B folds along a polarized N-terminal pathway.Structure. 2008 May;16(5):705-14. doi: 10.1016/j.str.2008.02.015. Structure. 2008. PMID: 18462675 Free PMC article.
References
-
- Plaxco KW, Simons KT, Ruczinski I, Baker D. Topology, stability, sequence, and length: defining the determinants of two-state protein folding kinetics. Biochemistry. 2000;39:11177–11183. - PubMed
-
- Sanchez IE, Kiefhaber T. Evidence for sequential barriers and obligatory intermediates in apparent two-state protein folding. J Mol Biol. 2003;325:367–376. - PubMed
-
- Sanchez IE, Kiefhaber T. Origin of unusual phi-values in protein folding: evidence against specific nucleation sites. J Mol Biol. 2003;334:1077–1085. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

