Smad4 is required to regulate the fate of cranial neural crest cells
- PMID: 17964566
- PMCID: PMC2704603
- DOI: 10.1016/j.ydbio.2007.09.050
Smad4 is required to regulate the fate of cranial neural crest cells
Abstract
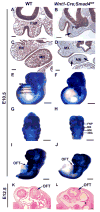
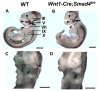
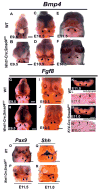
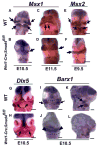
Smad4 is the central mediator for TGF-beta/BMP signals, which are involved in regulating cranial neural crest (CNC) cell formation, migration, proliferation and fate determination. It is unclear whether TGF-beta/BMP signals utilize Smad-dependent or -independent pathways to control the development of CNC cells. To investigate the functional significance of Smad4 in regulating CNC cells, we generated mice with neural crest specific inactivation of the Smad4 gene. Our study shows that Smad4 is not required for the migration of CNC cells, but is required in neural crest cells for the development of the cardiac outflow tract. Smad4 is essential in mediating BMP signaling in the CNC-derived ectomesenchyme during early stages of tooth development because conditional inactivation of Smad4 in neural crest derived cells results in incisor and molar development arrested at the dental lamina stage. Furthermore, Smad-mediated TGF-beta/BMP signaling controls the homeobox gene patterning of oral/aboral and proximal/distal domains within the first branchial arch. At the cellular level, a Smad4-mediated downstream target gene(s) is required for the survival of CNC cells in the proximal domain of the first branchial arch. Smad4 mutant mice show underdevelopment of the first branchial arch and midline fusion defects. Taken together, our data show that TGF-beta/BMP signals rely on Smad-dependent pathways in the ectomesenchyme to mediate epithelial-mesenchymal interactions that control craniofacial organogenesis.
Figures




Similar articles
-
SMAD4-mediated WNT signaling controls the fate of cranial neural crest cells during tooth morphogenesis.Development. 2011 May;138(10):1977-89. doi: 10.1242/dev.061341. Epub 2011 Apr 13. Development. 2011. PMID: 21490069 Free PMC article.
-
TGF-beta type I receptor Alk5 regulates tooth initiation and mandible patterning in a type II receptor-independent manner.Dev Biol. 2008 Aug 1;320(1):19-29. doi: 10.1016/j.ydbio.2008.03.045. Epub 2008 Apr 15. Dev Biol. 2008. PMID: 18572160 Free PMC article.
-
Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects.Development. 2003 Nov;130(21):5269-80. doi: 10.1242/dev.00708. Development. 2003. PMID: 12975342
-
Gene regulatory network from cranial neural crest cells to osteoblast differentiation and calvarial bone development.Cell Mol Life Sci. 2022 Feb 27;79(3):158. doi: 10.1007/s00018-022-04208-2. Cell Mol Life Sci. 2022. PMID: 35220463 Free PMC article. Review.
-
Tooth and jaw: molecular mechanisms of patterning in the first branchial arch.Arch Oral Biol. 2003 Jan;48(1):1-14. doi: 10.1016/s0003-9969(02)00208-x. Arch Oral Biol. 2003. PMID: 12615136 Review.
Cited by
-
Cleft palate defect of Dlx1/2-/- mutant mice is caused by lack of vertical outgrowth in the posterior palate.Dev Dyn. 2012 Nov;241(11):1757-69. doi: 10.1002/dvdy.23867. Epub 2012 Sep 28. Dev Dyn. 2012. PMID: 22972697 Free PMC article.
-
Abnormal dental follicle cells: A crucial determinant in tooth eruption disorders (Review).Mol Med Rep. 2024 Sep;30(3):168. doi: 10.3892/mmr.2024.13292. Epub 2024 Jul 19. Mol Med Rep. 2024. PMID: 39027997 Free PMC article. Review.
-
Inactivation of Tgfbr2 in Osterix-Cre expressing dental mesenchyme disrupts molar root formation.Dev Biol. 2013 Oct 1;382(1):27-37. doi: 10.1016/j.ydbio.2013.08.003. Epub 2013 Aug 8. Dev Biol. 2013. PMID: 23933490 Free PMC article.
-
BMP Signaling in the Development and Regeneration of Cranium Bones and Maintenance of Calvarial Stem Cells.Front Cell Dev Biol. 2020 Mar 10;8:135. doi: 10.3389/fcell.2020.00135. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32211409 Free PMC article. Review.
-
Homozygous missense N629D hERG (KCNH2) potassium channel mutation causes developmental defects in the right ventricle and its outflow tract and embryonic lethality.Circ Res. 2008 Dec 5;103(12):1483-91. doi: 10.1161/CIRCRESAHA.108.177055. Epub 2008 Oct 23. Circ Res. 2008. PMID: 18948620 Free PMC article.
References
-
- Allen SP, Borgadi JP, Barlow AJ, Mir SA, Qayyum SR, Verbeek FJ, Anderson RH, Francis-West PH, Brown NA, Richardson MK. Misexpression of noggin leads to septal defects in the outflow tract of the chick heart. Dev Biol. 2001;235:98–109. - PubMed
-
- Baker CV, Bronner-Fraser M. Vertebrate cranial placodes I. Embryonic induction. Dev Biol. 2001;232:1–61. - PubMed
-
- Barlow LA. Cranial nerve development: placodal neurons ride the crest. Curr Biol. 2002;12:R171–R173. - PubMed
-
- Beverdam A, Brouwer A, Reijnen M, Korving J, Meijlink F. Severe nasal clefting and abnormal embryonic apoptosis in Alx3/Alx4 double mutant mice. Development. 2001;128:3975–3986. - PubMed
-
- Chai Y, Jiang X, Ito Y, Bringas P, Jr, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

