Genomic analysis of human microRNA transcripts
- PMID: 17965236
- PMCID: PMC2077053
- DOI: 10.1073/pnas.0703890104
Genomic analysis of human microRNA transcripts
Abstract
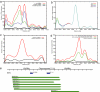
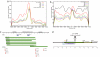
MicroRNAs (miRNAs) are important genetic regulators of development, differentiation, growth, and metabolism. The mammalian genome encodes approximately 500 known miRNA genes. Approximately 50% are expressed from non-protein-coding transcripts, whereas the rest are located mostly in the introns of coding genes. Intronic miRNAs are generally transcribed coincidentally with their host genes. However, the nature of the primary transcript of intergenic miRNAs is largely unknown. We have performed a large-scale analysis of transcription start sites, polyadenylation signals, CpG islands, EST data, transcription factor-binding sites, and expression ditag data surrounding intergenic miRNAs in the human genome to improve our understanding of the structure of their primary transcripts. We show that a significant fraction of primary transcripts of intergenic miRNAs are 3-4 kb in length, with clearly defined 5' and 3' boundaries. We provide strong evidence for the complete transcript structure of a small number of human miRNAs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Bartel DP. Cell. 2004;116:281–297. - PubMed
-
- Pasquinelli AE, Hunter S, Bracht J. Curr Opin Genet Dev. 2005;15:200–205. - PubMed
-
- Wightman B, Ha I, Ruvkun G. Cell. 1993;75:855–862. - PubMed
-
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Science. 2006;312:75–79. - PubMed
-
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Cell. 2003;115:787–798. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

