Lossless compression of chemical fingerprints using integer entropy codes improves storage and retrieval
- PMID: 17967006
- PMCID: PMC2536658
- DOI: 10.1021/ci700200n
Lossless compression of chemical fingerprints using integer entropy codes improves storage and retrieval
Abstract
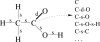
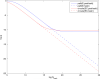
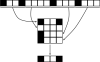
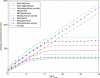
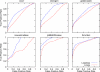
Many modern chemoinformatics systems for small molecules rely on large fingerprint vector representations, where the components of the vector record the presence or number of occurrences in the molecular graphs of particular combinatorial features, such as labeled paths or labeled trees. These large fingerprint vectors are often compressed to much shorter fingerprint vectors using a lossy compression scheme based on a simple modulo procedure. Here, we combine statistical models of fingerprints with integer entropy codes, such as Golomb and Elias codes, to encode the indices or the run lengths of the fingerprints. After reordering the fingerprint components by decreasing frequency order, the indices are monotone-increasing and the run lengths are quasi-monotone-increasing, and both exhibit power-law distribution trends. We take advantage of these statistical properties to derive new efficient, lossless, compression algorithms for monotone integer sequences: monotone value (MOV) coding and monotone length (MOL) coding. In contrast to lossy systems that use 1024 or more bits of storage per molecule, we can achieve lossless compression of long chemical fingerprints based on circular substructures in slightly over 300 bits per molecule, close to the Shannon entropy limit, using a MOL Elias Gamma code for run lengths. The improvement in storage comes at a modest computational cost. Furthermore, because the compression is lossless, uncompressed similarity (e.g., Tanimoto) between molecules can be computed exactly from their compressed representations, leading to significant improvements in retrival performance, as shown on six benchmark data sets of druglike molecules.
Figures




References
-
- Fligner MA, Verducci JS, Blower PE. A Modification of the Jaccard/Tanimoto Similarity Index for Diverse Selection of Chemical Compounds Using Binary Strings. Technometrics. 2002;44:110–119.
-
- Flower DR. On the properties of bit string-based measures of chemical similarity. Journal of Chemical Information and Computer Science. 1998;38:378–386.
-
- James CA, Weininger D, Delany J. Daylight Theory Manual. 2004. Current 2007 version available at http://www.daylight.com/dayhtml/doc/theory/index.html.
-
- Xue L, Godden JF, Stahura FL, Bajorath J. Profile scaling increases the similarity search performance of molecular fingerprints containing numerical descriptors and structural keys. Journal of Chemical Information and Computer Sciences. 2003;43:1218–1225. - PubMed
-
- Xue L, Stahura FL, Bajorath J. Similarity search profiling reveals effects of fingerprint scaling in virtual screening. Journal of Chemical Information and Computer Sciences. 2004;44:2032–2039. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

