Target cell APOBEC3C can induce limited G-to-A mutation in HIV-1
- PMID: 17967058
- PMCID: PMC2042017
- DOI: 10.1371/journal.ppat.0030153
Target cell APOBEC3C can induce limited G-to-A mutation in HIV-1
Abstract
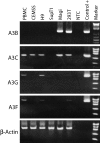
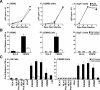
The evolutionary success of primate lentiviruses reflects their high capacity to mutate and adapt to new host species, immune responses within individual hosts, and, in recent years, antiviral drugs. APOBEC3G (A3G) and APOBEC3F (A3F) are host cell DNA-editing enzymes that induce extensive HIV-1 mutation that severely attenuates viral replication. The HIV-1 virion infectivity factor (Vif), expressed in vivo, counteracts the antiviral activity of A3G and A3F by inducing their degradation. Other APOBECs may contribute more to viral diversity by inducing less extensive mutations allowing viral replication to persist. Here we show that in APOBEC3C (A3C)-expressing cells infected with the patient-derived HIV-1 molecular clones 210WW, 210WM, 210MW, and 210MM, and the lab-adapted molecular clone LAI, viral G-to-A mutations were detected in the presence of Vif expression. Mutations occurred primarily in the GA context and were relatively infrequent, thereby allowing for spreading infection. The mutations were absent in cells lacking A3C but were induced after transient expression of A3C in the infected target cell. Inhibiting endogenous A3C by RNA interference in Magi cells prevented the viral mutations. Thus, A3C is necessary and sufficient for G-to-A mutations in some HIV-1 strains. A3C-induced mutations occur at levels that allow replication to persist and may therefore contribute to viral diversity. Developing drugs that inhibit A3C may be a novel strategy for delaying viral escape from immune or antiretroviral inhibition.
Conflict of interest statement
Figures

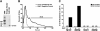
References
-
- Chen Z, Telfier P, Gettie A, Reed P, Zhang L, et al. Genetic characterization of new West African simian immunodeficiency virus SIVsm: Geographic clustering of household-derived SIV strains with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop. J Virol. 1996;70:3617–3627. - PMC - PubMed
-
- Hahn BH, Shaw GM, De Cock KM, Sharp PM. AIDS as a zoonosis: Scientific and public health implications. Science. 2000;287:607–614. - PubMed
-
- Phillips RE, Rowland-Jones S, Nixon DF, Gotch FM, Edwards JP, et al. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature. 1991;354:453–459. - PubMed
-
- Fitzgibbon JE, Mazar S, Dubin DT. A new type of G–>A hypermutation affecting human immunodeficiency virus. AIDS Res Hum Retroviruses. 1993;9:833–838. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

