Small-molecule inhibition of HIV pre-mRNA splicing as a novel antiretroviral therapy to overcome drug resistance
- PMID: 17967062
- PMCID: PMC2042022
- DOI: 10.1371/journal.ppat.0030159
Small-molecule inhibition of HIV pre-mRNA splicing as a novel antiretroviral therapy to overcome drug resistance
Abstract
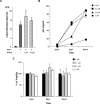
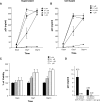
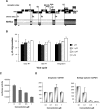
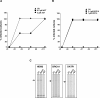
The development of multidrug-resistant viruses compromises antiretroviral therapy efficacy and limits therapeutic options. Therefore, it is an ongoing task to identify new targets for antiretroviral therapy and to develop new drugs. Here, we show that an indole derivative (IDC16) that interferes with exonic splicing enhancer activity of the SR protein splicing factor SF2/ASF suppresses the production of key viral proteins, thereby compromising subsequent synthesis of full-length HIV-1 pre-mRNA and assembly of infectious particles. IDC16 inhibits replication of macrophage- and T cell-tropic laboratory strains, clinical isolates, and strains with high-level resistance to inhibitors of viral protease and reverse transcriptase. Importantly, drug treatment of primary blood cells did not alter splicing profiles of endogenous genes involved in cell cycle transition and apoptosis. Thus, human splicing factors represent novel and promising drug targets for the development of antiretroviral therapies, particularly for the inhibition of multidrug-resistant viruses.
Conflict of interest statement
Figures


References
-
- Geretti AM. Clinical implications of HIV drug resistance to nucleoside and nucleotide reverse transcriptase inhibitors. AIDS Rev. 2006;8:210–220. - PubMed
-
- Del Rio C. Current concepts in antiretroviral therapy failure. Top HIV Med. 2006;14:102–106. - PubMed
-
- Barbaro G, Scozzafava A, Mastrolorenzo A, Supuran CT. Highly active antiretroviral therapy: current state of the art, new agents and their pharmacological interactions useful for improving therapeutic outcome. Curr Pharm Des. 2005;11:1805–1843. - PubMed
-
- Kijak GH, Currier JR, Tovanabutra S, Cox JH, Michael NL, et al. Lost in translation: implications of HIV-1 codon usage for immune escape and drug resistance. AIDS Rev. 2004;6:54–60. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

