Role of APOBEC3 in genetic diversity among endogenous murine leukemia viruses
- PMID: 17967065
- PMCID: PMC2041998
- DOI: 10.1371/journal.pgen.0030183
Role of APOBEC3 in genetic diversity among endogenous murine leukemia viruses
Abstract
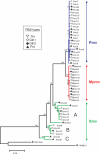
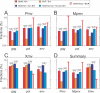
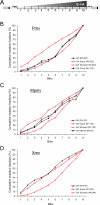
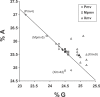
The ability of human and murine APOBECs (specifically, APOBEC3) to inhibit infecting retroviruses and retrotransposition of some mobile elements is becoming established. Less clear is the effect that they have had on the establishment of the endogenous proviruses resident in the human and mouse genomes. We used the mouse genome sequence to study diversity and genetic traits of nonecotropic murine leukemia viruses (polytropic [Pmv], modified polytropic [Mpmv], and xenotropic [Xmv] subgroups), the best-characterized large set of recently integrated proviruses. We identified 49 proviruses. In phylogenetic analyses, Pmvs and Mpmvs were monophyletic, whereas Xmvs were divided into several clades, implying a greater number of replication cycles between the integration events. Four distinct primer binding site types (Pro, Gln1, Gln2 and Thr) were dispersed within the phylogeny, indicating frequent mispriming. We analyzed the frequency and context of G-to-A mutations for the role of mA3 in formation of these proviruses. In the Pmv and Mpmv (but not Xmv) groups, mutations attributable to mA3 constituted a large fraction of the total. A significant number of nonsense mutations suggests the absence of purifying selection following mutation. A strong bias of G-to-A relative to C-to-T changes was seen, implying a strand specificity that can only have occurred prior to integration. The optimal sequence context of G-to-A mutations, TTC, was consistent with mA3. At least in the Pmv group, a significant 5' to 3' gradient of G-to-A mutations was consistent with mA3 editing. Altogether, our results for the first time suggest mA3 editing immediately preceding the integration event that led to retroviral endogenization, contributing to inactivation of infectivity.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures

References
-
- Boeke JD, Stoye JP. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. New York: Cold Spring Harbor Laboratory Press; 1997. pp. 343–436. - PubMed
-
- CSAC. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. - PubMed
-
- International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. - PubMed
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Telesnitsky A, Goff SP. Reverse transcriptase and the generation of retroviral DNA. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. New York: Cold Spring Harbor Laboratory Press; 1997. pp. 121–160. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

