Model systems for understanding DNA base pairing
- PMID: 17967435
- PMCID: PMC2175026
- DOI: 10.1016/j.cbpa.2007.09.019
Model systems for understanding DNA base pairing
Abstract
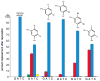
The fact that nucleic acid bases recognize each other to form pairs is a canonical part of the dogma of biology. However, they do not recognize each other well enough in water to account for the selectivity and efficiency that is needed in the transmission of biological information through a cell. Thus proteins assist in this recognition in multiple ways, and recent data suggest that these mechanisms of recognition can vary widely with context. To probe how the chemical differences of the four nucleobases are defined in various biological contexts, chemists and biochemists have developed modified versions that differ in their polarity, shape, size, and functional groups. This brief review covers recent advances in this field of research.
Figures
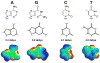

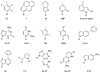
Similar articles
-
Measurement and theory of hydrogen bonding contribution to isosteric DNA base pairs.J Am Chem Soc. 2012 Feb 15;134(6):3154-63. doi: 10.1021/ja210475a. Epub 2012 Feb 2. J Am Chem Soc. 2012. PMID: 22300089 Free PMC article.
-
Metal-mediated DNA base pairing: alternatives to hydrogen-bonded Watson-Crick base pairs.Acc Chem Res. 2012 Dec 18;45(12):2066-76. doi: 10.1021/ar200313h. Epub 2012 Mar 27. Acc Chem Res. 2012. PMID: 22452649
-
Sequence recognition in the pairing of DNA duplexes.Phys Rev Lett. 2001 Apr 16;86(16):3666-9. doi: 10.1103/PhysRevLett.86.3666. Phys Rev Lett. 2001. PMID: 11328049
-
The steric hypothesis for DNA replication and fluorine hydrogen bonding revisited in light of structural data.Acc Chem Res. 2012 Aug 21;45(8):1237-46. doi: 10.1021/ar200303k. Epub 2012 Apr 23. Acc Chem Res. 2012. PMID: 22524491 Free PMC article. Review.
-
Unnatural nucleosides with unusual base pairing properties.Curr Protoc Nucleic Acid Chem. 2001 Aug;Chapter 1:Unit 1.4. doi: 10.1002/0471142700.nc0104s05. Curr Protoc Nucleic Acid Chem. 2001. PMID: 18428819 Review.
Cited by
-
Templating efficiency of naked DNA.Proc Natl Acad Sci U S A. 2010 Jul 6;107(27):12074-9. doi: 10.1073/pnas.0914872107. Epub 2010 Jun 16. Proc Natl Acad Sci U S A. 2010. PMID: 20554916 Free PMC article.
-
DNA interstrand cross-linking upon irradiation of aryl halide C-nucleotides.J Org Chem. 2014 Mar 7;79(5):1877-84. doi: 10.1021/jo4028227. Epub 2014 Feb 24. J Org Chem. 2014. PMID: 24559326 Free PMC article.
-
Non-Covalent Forces in Naphthazarin-Cooperativity or Competition in the Light of Theoretical Approaches.Int J Mol Sci. 2021 Jul 27;22(15):8033. doi: 10.3390/ijms22158033. Int J Mol Sci. 2021. PMID: 34360798 Free PMC article.
-
Bromopyridone Nucleotide Analogues, Anoxic Selective Radiosensitizing Agents That Are Incorporated in DNA by Polymerases.J Org Chem. 2015 Nov 6;80(21):10675-85. doi: 10.1021/acs.joc.5b01833. Epub 2015 Oct 28. J Org Chem. 2015. PMID: 26509218 Free PMC article.
-
Detection of bacterial 16S rRNA using a molecular beacon-based X sensor.Biosens Bioelectron. 2013 Mar 15;41:386-90. doi: 10.1016/j.bios.2012.08.058. Epub 2012 Sep 5. Biosens Bioelectron. 2013. PMID: 23021850 Free PMC article.
References
-
- Kim TW, Kool ET. A series of nonpolar thymine analogues of increasing size: DNA base pairing and stacking properties. J Org Chem. 2005;70:2048–2053. - PubMed
-
- Kim TW, Brieba LG, Ellenberger T, Kool ET. Functional evidence for a small and rigid active site in a high fidelity DNA polymerase: probing T7 DNA polymerase with variably sized base pairs. J Biol Chem. 2006;281:2289–2295. - PubMed
-
- Hirao I, Kimoto M, Mitsui T, Fujiwara T, Kawai R, Sato A, Harada Y, Yokoyama S. An unnatural hydrophobic base pair system: site-specific incorporation of nucleotide analogs into DNA and RNA. Nat Methods. 2006;9:729–735. - PubMed
-
- Mitsui T, Kimoto M, Harada Y, Yokoyama S, Hirao I. An efficient unnatural base pair for a base-pair-expanded transcription system. J Am Chem Soc. 2005;127:8652–8658. A particularly effective example of a nonnatural, non-hydrogen-bonded base pair that is a substrate for polymerases. It is has been shown to have useful applications in labeling RNAs. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

