Regulation of Notch signaling by dynamic changes in the precision of S3 cleavage of Notch-1
- PMID: 17967888
- PMCID: PMC2223315
- DOI: 10.1128/MCB.00863-07
Regulation of Notch signaling by dynamic changes in the precision of S3 cleavage of Notch-1
Abstract
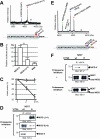
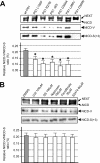
Intramembrane proteolysis by presenilin-dependent gamma-secretase produces the Notch intracellular cytoplasmic domain (NCID) and Alzheimer disease-associated amyloid-beta. Here, we show that upon Notch signaling the intracellular domain of Notch-1 is cleaved into two distinct types of NICD species due to diversity in the site of S3 cleavage. Consistent with the N-end rule, the S3-V cleavage produces stable NICD with Val at the N terminus, whereas the S3-S/S3-L cleavage generates unstable NICD with Ser/Leu at the N terminus. Moreover, intracellular Notch signal transmission with unstable NICDs is much weaker than that with stable NICD. Importantly, the extent of endocytosis in target cells affects the relative production ratio of the two types of NICD, which changes in parallel with Notch signaling. Surprisingly, substantial amounts of unstable NICD species are generated from the Val-->Gly and the Lys-->Arg mutants, which have been reported to decrease S3 cleavage efficiency in cultured cells. Thus, we suggest that the existence of two distinct types of NICD points to a novel aspect of the intracellular signaling and that changes in the precision of S3 cleavage play an important role in the process of conversion from extracellular to intracellular Notch signaling.
Figures



Similar articles
-
Role of the RAM domain and ankyrin repeats on notch signaling and activity in cells of osteoblastic lineage.J Bone Miner Res. 2006 Aug;21(8):1317-26. doi: 10.1359/jbmr.060505. J Bone Miner Res. 2006. PMID: 16869730
-
Quantification of gamma-secretase modulation differentiates inhibitor compound selectivity between two substrates Notch and amyloid precursor protein.Mol Brain. 2008 Nov 4;1:15. doi: 10.1186/1756-6606-1-15. Mol Brain. 2008. PMID: 18983676 Free PMC article.
-
[Amyloid-beta 42 generating process may have a biological role in regulation of Notch signaling intensity].Nihon Shinkei Seishin Yakurigaku Zasshi. 2008 Nov;28(5-6):177-83. Nihon Shinkei Seishin Yakurigaku Zasshi. 2008. PMID: 19108503 Review. Japanese.
-
Notch signaling proteins HES-1 and Hey-1 bind to insulin degrading enzyme (IDE) proximal promoter and repress its transcription and activity: implications for cellular Aβ metabolism.Biochim Biophys Acta. 2012 Feb;1823(2):227-35. doi: 10.1016/j.bbamcr.2011.09.014. Epub 2011 Oct 19. Biochim Biophys Acta. 2012. PMID: 22036964 Free PMC article.
-
Processes of beta-amyloid and intracellular cytoplasmic domain generation by presenilin/gamma-secretase.Neurodegener Dis. 2008;5(3-4):160-2. doi: 10.1159/000113690. Epub 2008 Mar 6. Neurodegener Dis. 2008. PMID: 18322378 Review.
Cited by
-
Misoprostol-Induced Modification of the Notch Signaling Pathway in the Human Cervix.Reprod Sci. 2019 Jul;26(7):909-917. doi: 10.1177/1933719118799208. Epub 2018 Oct 3. Reprod Sci. 2019. PMID: 30278829
-
The nuclear pore proteins Nup88/214 and T-cell acute lymphatic leukemia-associated NUP214 fusion proteins regulate Notch signaling.J Biol Chem. 2019 Aug 2;294(31):11741-11750. doi: 10.1074/jbc.RA118.006357. Epub 2019 Jun 11. J Biol Chem. 2019. PMID: 31186352 Free PMC article.
-
Competition and collaboration: GATA-3, PU.1, and Notch signaling in early T-cell fate determination.Semin Immunol. 2008 Aug;20(4):236-46. doi: 10.1016/j.smim.2008.07.006. Epub 2008 Sep 3. Semin Immunol. 2008. PMID: 18768329 Free PMC article. Review.
-
Class II phosphatidylinositol 3-kinase-C2α is essential for Notch signaling by regulating the endocytosis of γ-secretase in endothelial cells.Sci Rep. 2021 Mar 4;11(1):5199. doi: 10.1038/s41598-021-84548-4. Sci Rep. 2021. PMID: 33664344 Free PMC article.
-
Notch signaling from the endosome requires a conserved dileucine motif.Mol Biol Cell. 2013 Feb;24(3):297-307. doi: 10.1091/mbc.E12-02-0081. Epub 2012 Nov 21. Mol Biol Cell. 2013. PMID: 23171551 Free PMC article.
References
-
- Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science 284770-776. - PubMed
-
- Bachmair, A., D. Finley, and A. Varshavsky. 1986. In vivo half-life of a protein is a function of its amino-terminal residue. Science 234179-186. - PubMed
-
- Berdnik, D., T. Torok, M. Gonzalez-Gaitan, and J. A. Knoblich. 2002. The endocytic protein alpha-adaptin is required for numb-mediated asymmetric cell division in Drosophila. Dev. Cell 3221-231. - PubMed
-
- Blat, Y., J. E. Meredith, Q. Wang, J. D. Bradley, L. A. Thompson, R. E. Olson, A. M. Stern, and D. Seiffert. 2002. Mutations at the P1′ position of Notch1 decrease intracellular domain stability rather than cleavage by gamma-secretase. Biochem. Biophys. Res. Commun. 299569-573. - PubMed
-
- Cao, X., and T. C. Sudhof. 2001. A transcriptionally active complex of APP with Fe65 and histone acetyltransferase Tip60. Science 293115-120. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
