A novel quantitative trait locus on mouse chromosome 18, "era1," modifies the entrainment of circadian rhythms
- PMID: 17969459
- PMCID: PMC2266285
- DOI: 10.1093/sleep/30.10.1255
A novel quantitative trait locus on mouse chromosome 18, "era1," modifies the entrainment of circadian rhythms
Abstract
Study objective: The mammalian circadian clock in the suprachiasmatic nuclei (SCN) of the hypothalamus conveys 24-h rhythmicity to sleep-wake cycles, locomotor activity, and other behavioral and physiological processes. The timing of rhythms relative to the light/dark (LD12:12) cycle is influenced in part by the endogenous circadian period and the time of day specific sensitivity of the clock to light. We now describe a novel circadian rhythm phenotype, and a locus influencing that phenotype, in a segregating population of mice.
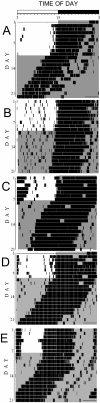
Methods: By crossbreeding 2 genetically distinct nocturnal strains of mice (Cast/Ei and C57BL/6J) and backcrossing the resulting progeny to Cast/Ei, we have produced a novel circadian phenotype, called early runner mice.
Results: Early runner mice entrain to a light/dark cycle at an advanced phase, up to 9 hours before dark onset. This phenotype is not significantly correlated with circadian period in constant darkness and is not associated with disruption of molecular circadian rhythms in the SCN, as assessed by analysis of period gene expression. We have identified a genomic region that regulates this phenotype-a major quantitative trait locus on chromosome 18 (near D18Mit184) that we have named era1 for Early Runner Activity locus one. Phase delays caused by light exposure early in the subjective night were of smaller magnitude in backcross offspring that were homozygous Cast/Ei at D18Mit184 than in those that were heterozygous at this locus.
Conclusion: Genetic variability in the circadian response to light may, in part, explain the variance in phase angle of entrainment in this segregating mouse population.
Figures






References
-
- Edgar DM. Functional role of the suprachiasmatic nuclei in the regulation of sleep and wakefulness. In: Guilleminault C, editor. Fatal familial insomnia: inherited prion diseases, sleep and the thalamus. New York: Raven Press, Ltd.; 1994. pp. 203–13.
-
- King DP, Takahashi JS. Molecular genetics of circadian rhythms in mammals. Annu Rev Neurosci. 2000;23:713–42. - PubMed
-
- Wisor JP. Disorders of the circadian clock: Etiology and possible therapeutic targets. Curr Drug Targets - CNS – Neurological Disorders. 2002;1:555–66. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

