Robustness and evolvability: a paradox resolved
- PMID: 17971325
- PMCID: PMC2562401
- DOI: 10.1098/rspb.2007.1137
Robustness and evolvability: a paradox resolved
Abstract
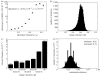
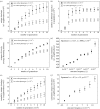
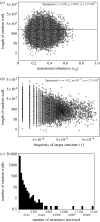
Understanding the relationship between robustness and evolvability is key to understand how living things can withstand mutations, while producing ample variation that leads to evolutionary innovations. Mutational robustness and evolvability, a system's ability to produce heritable variation, harbour a paradoxical tension. On one hand, high robustness implies low production of heritable phenotypic variation. On the other hand, both experimental and computational analyses of neutral networks indicate that robustness enhances evolvability. I here resolve this tension using RNA genotypes and their secondary structure phenotypes as a study system. To resolve the tension, one must distinguish between robustness of a genotype and a phenotype. I confirm that genotype (sequence) robustness and evolvability share an antagonistic relationship. In stark contrast, phenotype (structure) robustness promotes structure evolvability. A consequence is that finite populations of sequences with a robust phenotype can access large amounts of phenotypic variation while spreading through a neutral network. Population-level processes and phenotypes rather than individual sequences are key to understand the relationship between robustness and evolvability. My observations may apply to other genetic systems where many connected genotypes produce the same phenotypes.
Figures

References
-
- Aharoni A, Gaidukov L, Khersonsky O, Gould S.M, Roodveldt C, Tawfik D.S. The ‘evolvability’ of promiscuous protein functions. Nat. Genet. 2005;37:73–76. - PubMed
-
- Ancel L.W, Fontana W. Plasticity, evolvability, and modularity in RNA. J. Exp. Zool./Mol. Dev. Evol. 2000;288:242–283. doi:10.1002/1097-010X(20001015)288:3<242::AID-JEZ5>3.0.CO;2-O - DOI - PubMed
-
- Baudin F, Marquet R, Isel C, Darlix J, Ehresmann B, Ehresmann C. Functional sites in the 5′ region of human-immunodeficiency-virus type-1 RNA form defined structural domains. J. Mol. Biol. 1993;229:382–397. doi:10.1006/jmbi.1993.1041 - DOI - PubMed
-
- Bedau M.A, Packard N.H. Evolution of evolvability via adaptation of mutation rates. Biosystems. 2003;69:143–162. doi:10.1016/S0303-2647(02)00137-5 - DOI - PubMed
-
- Bloom J.D, Labthavikul S.T, Otey C.R, Arnold F.H. Protein stability promotes evolvability. Proc. Natl Acad. Sci. USA. 2006;103:5869–5874. doi:10.1073/pnas.0510098103 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

