Evidence for positive selection in the C-terminal domain of the cholesterol metabolism gene PCSK9 based on phylogenetic analysis in 14 primate species
- PMID: 17971861
- PMCID: PMC2034530
- DOI: 10.1371/journal.pone.0001098
Evidence for positive selection in the C-terminal domain of the cholesterol metabolism gene PCSK9 based on phylogenetic analysis in 14 primate species
Abstract
Background: Cholesterol homeostasis is maintained through finely tuned mechanisms regulating intestinal absorption, hepatic biosynthesis and secretion as well as plasma clearance. Proprotein convertase subtilisin/kexin type 9 (PCSK9) is a secreted enzyme of the serine protease family that reduces cellular uptake of plasma low-density lipoprotein (LDL) cholesterol by promoting LDL receptor (LDL-R) degradation. Species-specific positive selection has been noted in the LDLR promoter, leading to differential expression of LDLR among primates. Whether PCSK9 experienced significant selective pressure to maintain a functional relationship with its target protein, LDL-R, is unknown.
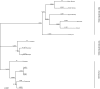
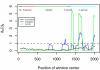
Methodology/principal findings: We compiled the sequences of the coding regions of PCSK9 from 14 primate species in the clade of Hominoids, Old World monkeys and New World monkeys. To detect selective pressure at the protein level, the ratios of nonsynonymous/synonymous substitution rate (d(N)/d(S)) under different evolutionary models were calculated across the phylogeny of PCSK9. Maximum likelihood analyses of d(N)/d(S) ratios for the aligned coding region sequences among 14 primate species indicated that PCSK9 was subject to a strong functional constraint (i.e., purifying selection). However, positive selection was noted in the functional carboxyl-terminal (C-terminal) domain in many branches across the phylogeny, especially in the lineage leading to the orangutan. Furthermore, at least five positively selected amino acids were detected in this lineage using the branch-site model A. In a sliding-window analysis, several d(N)/d(S) peaks in the C-terminal domain in both the human and the orangutan branches were noted.
Conclusions: These results suggest that among primates, differential selective pressure has shaped evolutionary patterns in the functional domains of PCSK9, an important regulator of cholesterol homeostasis.
Conflict of interest statement
Figures



References
-
- Espenshade PJ, Cheng D, Goldstein JL, Brown MS. Autocatalytic processing of site-1 protease removes propeptide and permits cleavage of sterol regulatory element-binding proteins. J Biol Chem. 1999;274:22795–22804. - PubMed
-
- Elagoz A, Benjannet S, Mammarbassi A, Wickham L, Seidah NG. Biosynthesis and cellular trafficking of the convertase SKI-1/S1P: ectodomain shedding requires SKI-1 activity. J Biol Chem. 2002;277:11265–11275. - PubMed
-
- Benjannet S, Rhainds D, Essalmani R, Mayne J, Wickham L, et al. NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J Biol Chem. 2004;279:48865–48875. - PubMed
-
- Naureckiene S, Ma L, Sreekumar K, Purandare U, Lo CF, et al. Functional characterization of Narc 1, a novel proteinase related to proteinase K. Arch Biochem Biophys. 2003;420:55–67. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

