Molecular correlates of host specialization in Staphylococcus aureus
- PMID: 17971880
- PMCID: PMC2040198
- DOI: 10.1371/journal.pone.0001120
Molecular correlates of host specialization in Staphylococcus aureus
Abstract
Background: The majority of Staphylococcus aureus isolates that are recovered from either serious infections in humans or from mastitis in cattle represent genetically distinct sets of clonal groups. Moreover, population genetic analyses have provided strong evidence of host specialization among S. aureus clonal groups associated with human and ruminant infection. However, the molecular basis of host specialization in S. aureus is not understood.
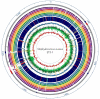
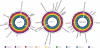
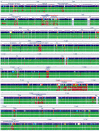
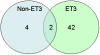
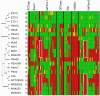
Methodology/principal findings: We sequenced the genome of strain ET3-1, a representative isolate of a common bovine mastitis-causing S. aureus clone. Strain ET3-1 encodes several genomic elements that have not been previously identified in S. aureus, including homologs of virulence factors from other gram-positive pathogens. Relative to the other sequenced S. aureus associated with human infection, allelic variation in ET3-1 was high among virulence and surface-associated genes involved in host colonization, toxin production, iron metabolism, antibiotic resistance, and gene regulation. Interestingly, a number of well-characterized S. aureus virulence factors, including protein A and clumping factor A, exist as pseudogenes in ET3-1. Whole-genome DNA microarray hybridization revealed considerable similarity in the gene content of highly successful S. aureus clones associated with bovine mastitis, but not among those clones that are only infrequently recovered from bovine hosts.
Conclusions/significance: Whole genome sequencing and comparative genomic analyses revealed a set of molecular genetic features that distinguish clones of highly successful bovine-associated S. aureus optimized for mastitis pathogenesis in cattle from those that infect human hosts or are only infrequently recovered from bovine sources. Further, the results suggest that modern bovine specialist clones diverged from a common ancestor resembling human-associated S. aureus clones through a combination of foreign DNA acquisition and gene decay.
Conflict of interest statement
Figures


References
-
- Wells SJ, Ott SL, Seitzinger AH. Key health issues for dairy cattle–new and old. J Dairy Sci. 1998;81:3029–3035. - PubMed
-
- Anderson KL, Lyman RL, Bodeis-Jones SM, White DG. Genetic diversity and antimicrobial susceptibility profiles among mastitis-causing Staphylococcus aureus isolated from bovine milk samples. Am J Vet Res. 2006;67:1185–1191. - PubMed
-
- Reinoso EB, El-Sayed A, Lammler C, Bogni C, Zschock M. Genotyping of Staphylococcus aureus isolated from humans, bovine subclinical mastitis and food samples in Argentina. Microbiol Res 2006 - PubMed
-
- Cifrian E, Guidry AJ, O'Brien CN, Nickerson SC, Marquardt WW. Adherence of Staphylococcus aureus to cultured bovine mammary epithelial cells. J Dairy Sci. 1994;77:970–983. - PubMed
-
- Leitner G, Krifucks O, Glickman A, Younis A, Saran A. Staphylococcus aureus strains isolated from bovine mastitis: virulence, antibody production and protection from challenge in a mouse model. FEMS Immunol Med Microbiol. 2003;35:99–106. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

