Multifunctional class I transcription in Trypanosoma brucei depends on a novel protein complex
- PMID: 17972917
- PMCID: PMC2099468
- DOI: 10.1038/sj.emboj.7601905
Multifunctional class I transcription in Trypanosoma brucei depends on a novel protein complex
Abstract
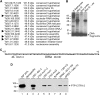
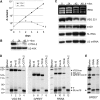
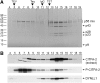
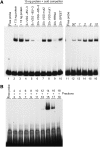
The vector-borne, protistan parasite Trypanosoma brucei is the only known eukaryote with a multifunctional RNA polymerase I that, in addition to ribosomal genes, transcribes genes encoding the parasite's major cell-surface proteins-the variant surface glycoprotein (VSG) and procyclin. In the mammalian bloodstream, antigenic variation of the VSG coat is the parasite's means to evade the immune response, while procyclin is necessary for effective establishment of trypanosome infection in the fly. Moreover, the exceptionally high efficiency of mono-allelic VSG expression is essential to bloodstream trypanosomes since its silencing caused rapid cell-cycle arrest in vitro and clearance of parasites from infected mice. Here we describe a novel protein complex that recognizes class I promoters and is indispensable for class I transcription; it consists of a dynein light chain and six polypeptides that are conserved only among trypanosomatid parasites. In accordance with an essential transcriptional function of the complex, silencing the expression of a key subunit was lethal to bloodstream trypanosomes and specifically affected the abundance of rRNA and VSG mRNA. The complex was dubbed class I transcription factor A.
Figures

References
-
- Grummt I (2003) Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev 17: 1691–1702 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

