Evaluation of the models handling heterotachy in phylogenetic inference
- PMID: 17974035
- PMCID: PMC2248194
- DOI: 10.1186/1471-2148-7-206
Evaluation of the models handling heterotachy in phylogenetic inference
Abstract
Background: The evolutionary rate at a given homologous position varies across time. When sufficiently pronounced, this phenomenon - called heterotachy - may produce artefactual phylogenetic reconstructions under the commonly used models of sequence evolution. These observations have motivated the development of models that explicitly recognize heterotachy, with research directions proposed along two main axes: 1) the covarion approach, where sites switch from variable to invariable states; and 2) the mixture of branch lengths (MBL) approach, where alignment patterns are assumed to arise from one of several sets of branch lengths, under a given phylogeny.
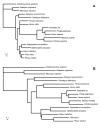
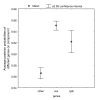
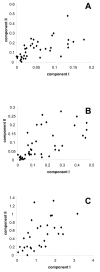
Results: Here, we report the first statistical comparisons contrasting the performance of covarion and MBL modeling strategies. Using simulations under heterotachous conditions, we explore the properties of three model comparison methods: the Akaike information criterion, the Bayesian information criterion, and cross validation. Although more time consuming, cross validation appears more reliable than AIC and BIC as it directly measures the predictive power of a model on 'future' data. We also analyze three large datasets (nuclear proteins of animals, mitochondrial proteins of mammals, and plastid proteins of plants), and find the optimal number of components of the MBL model to be two for all datasets, indicating that this model is preferred over the standard homogeneous model. However, the covarion model is always favored over the optimal MBL model.
Conclusion: We demonstrated, using three large datasets, that the covarion model is more efficient at handling heterotachy than the MBL model. This is probably due to the fact that the MBL model requires a serious increase in the number of parameters, as compared to two supplementary parameters of the covarion approach. Further improvements of the both the mixture and the covarion approaches might be obtained by modeling heterogeneous behavior both along time and across sites.
Figures

References
-
- Felsenstein J. Inferring phylogenies. Sunderland, MA, USA , Sinauer Associates, Inc.; 2004. p. 645.
-
- Yang Z. Maximum-likelihood estimation of phylogeny from DNA sequences when substitution rates differ over sites. Mol Biol Evol. 1993;10:1396–1401. - PubMed
-
- Lanave C, Preparata G, Saccone C, Serio G. A new method for calculating evolutionary substitution rates. J Mol Evol. 1984;20:86–93. - PubMed
-
- Galtier N. Maximum-likelihood phylogenetic analysis under a covarion-like model. Mol Biol Evol. 2001;18:866–873. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

