Structure of the retinoblastoma protein bound to adenovirus E1A reveals the molecular basis for viral oncoprotein inactivation of a tumor suppressor
- PMID: 17974914
- PMCID: PMC2045126
- DOI: 10.1101/gad.1590607
Structure of the retinoblastoma protein bound to adenovirus E1A reveals the molecular basis for viral oncoprotein inactivation of a tumor suppressor
Abstract
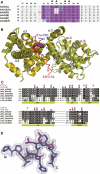
The adenovirus (Ad) E1A (Ad-E1A) oncoprotein mediates cell transformation, in part, by displacing E2F transcription factors from the retinoblastoma protein (pRb) tumor suppressor. In this study we determined the crystal structure of the pRb pocket domain in complex with conserved region 1 (CR1) of Ad5-E1A. The structure and accompanying biochemical studies reveal that E1A-CR1 binds at the interface of the A and B cyclin folds of the pRb pocket domain, and that both E1A-CR1 and the E2F transactivation domain use similar conserved nonpolar residues to engage overlapping sites on pRb, implicating a novel molecular mechanism for pRb inactivation by a viral oncoprotein.
Figures


References
-
- Arany Z., Newsome D., Oldread E., Livingston D.M., Eckner R., Newsome D., Oldread E., Livingston D.M., Eckner R., Oldread E., Livingston D.M., Eckner R., Livingston D.M., Eckner R., Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. - PubMed
-
- Avvakumov N., Kajon A.E., Hoeben R.C., Mymryk J.S., Kajon A.E., Hoeben R.C., Mymryk J.S., Hoeben R.C., Mymryk J.S., Mymryk J.S. Comprehensive sequence analysis of the E1A proteins of human and simian adenoviruses. Virology. 2004;329:477–492. - PubMed
-
- Boyer T.G., Martin M.E., Lees E., Ricciardi R.P., Berk A.J., Martin M.E., Lees E., Ricciardi R.P., Berk A.J., Lees E., Ricciardi R.P., Berk A.J., Ricciardi R.P., Berk A.J., Berk A.J. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature. 1999;399:276–279. - PubMed
-
- Dick F.A., Dyson N., Dyson N. pRB contains an E2F1-specific binding domain that allows E2F1-induced apoptosis to be regulated separately from other E2F activities. Mol. Cell. 2003;12:639–649. - PubMed
-
- Dyson N. The regulation of E2F by pRB-family proteins. Genes & Dev. 1998;12:2245–2262. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
