Initial sequence and comparative analysis of the cat genome
- PMID: 17975172
- PMCID: PMC2045150
- DOI: 10.1101/gr.6380007
Initial sequence and comparative analysis of the cat genome
Abstract
The genome sequence (1.9-fold coverage) of an inbred Abyssinian domestic cat was assembled, mapped, and annotated with a comparative approach that involved cross-reference to annotated genome assemblies of six mammals (human, chimpanzee, mouse, rat, dog, and cow). The results resolved chromosomal positions for 663,480 contigs, 20,285 putative feline gene orthologs, and 133,499 conserved sequence blocks (CSBs). Additional annotated features include repetitive elements, endogenous retroviral sequences, nuclear mitochondrial (numt) sequences, micro-RNAs, and evolutionary breakpoints that suggest historic balancing of translocation and inversion incidences in distinct mammalian lineages. Large numbers of single nucleotide polymorphisms (SNPs), deletion insertion polymorphisms (DIPs), and short tandem repeats (STRs), suitable for linkage or association studies were characterized in the context of long stretches of chromosome homozygosity. In spite of the light coverage capturing approximately 65% of euchromatin sequence from the cat genome, these comparative insights shed new light on the tempo and mode of gene/genome evolution in mammals, promise several research applications for the cat, and also illustrate that a comparative approach using more deeply covered mammals provides an informative, preliminary annotation of a light (1.9-fold) coverage mammal genome sequence.
Figures

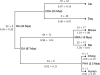
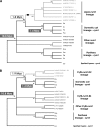




Comment in
-
2x genomes--does depth matter?Genome Res. 2007 Nov;17(11):1547-9. doi: 10.1101/gr.7050807. Genome Res. 2007. PMID: 17975171 No abstract available.
References
-
- Baker H., Smith B.R., Martin D.R., Foureman P., Smith B.R., Martin D.R., Foureman P., Martin D.R., Foureman P., Foureman P. Consultations in feline internal medicine. W.B. Saunders & Co.; Philadelphia: 2001.
-
- Batzoglou S., Jaffe D.B., Stanley K., Butler J., Gnerre S., Mauceli E., Berger B., Mesirov J.P., Lander E.S., Jaffe D.B., Stanley K., Butler J., Gnerre S., Mauceli E., Berger B., Mesirov J.P., Lander E.S., Stanley K., Butler J., Gnerre S., Mauceli E., Berger B., Mesirov J.P., Lander E.S., Butler J., Gnerre S., Mauceli E., Berger B., Mesirov J.P., Lander E.S., Gnerre S., Mauceli E., Berger B., Mesirov J.P., Lander E.S., Mauceli E., Berger B., Mesirov J.P., Lander E.S., Berger B., Mesirov J.P., Lander E.S., Mesirov J.P., Lander E.S., Lander E.S. ARACHNE: A whole-genome shotgun assembler. Genome Res. 2002;12:177–189. - PMC - PubMed
-
- Beck T.W., Menninger J., Murphy W.J., Nash W.G., O’Brien S.J., Yuhki N., Menninger J., Murphy W.J., Nash W.G., O’Brien S.J., Yuhki N., Murphy W.J., Nash W.G., O’Brien S.J., Yuhki N., Nash W.G., O’Brien S.J., Yuhki N., O’Brien S.J., Yuhki N., Yuhki N. The feline major histocompatibility complex is rearranged by an inversion with a breakpoint in the distal class I region. Immunogenetics. 2005;56:702–709. - PubMed
-
- Bejerano G., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D., Stephen S., Kent W.J., Mattick J.S., Haussler D., Kent W.J., Mattick J.S., Haussler D., Mattick J.S., Haussler D., Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. - PubMed
-
- Benveniste R.E., Lieber M.M., Livingston D.M., Sherr C.J., Todaro G.J., Kalter S.S., Lieber M.M., Livingston D.M., Sherr C.J., Todaro G.J., Kalter S.S., Livingston D.M., Sherr C.J., Todaro G.J., Kalter S.S., Sherr C.J., Todaro G.J., Kalter S.S., Todaro G.J., Kalter S.S., Kalter S.S. Infectious C-type virus isolated from a baboon placenta. Nature. 1974;248:17–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
