Detection and characterization of two ATP-dependent conformational changes in proteolytically inactive Escherichia coli Lon mutants by stopped flow kinetic techniques
- PMID: 17975895
- PMCID: PMC2537469
- DOI: 10.1021/bi701649b
Detection and characterization of two ATP-dependent conformational changes in proteolytically inactive Escherichia coli Lon mutants by stopped flow kinetic techniques
Abstract
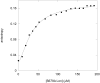
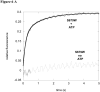
Lon is an ATP dependent serine protease responsible for degrading denatured, oxidatively damaged and certain regulatory proteins in the cell. In this study we exploited the fluorescence properties of a dansylated peptide substrate (S4) and the intrinsic Trp residues in Lon to monitor peptide interacting with the enzyme. We generated two proteolytically inactive Lon mutants, S679A and S679W, where the active site serine is mutated to an Ala and Trp residue, respectively. Stopped-flow fluorescence spectroscopy was used to identify key enzyme intermediates generated along the reaction pathway prior to peptide hydrolysis. A two-step peptide binding event is detected in both mutants, where a conformational change occurs after a rapid equilibrium peptide binding step. The Kd for the initial peptide binding step determined by kinetic and equilibrium binding techniques is approximately 164 micromolar and 38 micromolar, respectively. The rate constants for the conformational change detected in the S679A and S679W Lon mutants are 0.74 +/- 0.10 s(-1) and 0.57 +/- 0.10 s(-1), respectively. These values are comparable to the lag rate constant determined for peptide hydrolysis (klag approximately 1 s(-1)) [Vineyard, D., et al. (2005) Biochemistry 45, 4602-4610]. Replacement of the active site Ser with Trp (S679W) allows for the detection of an ATP-dependent conformational change within the proteolytic site. The rate constant for this conformational change is 7.6 +/- 1.0 s(-1), and is essentially identical to the burst rate constant determined for ATP hydrolysis under comparable reaction conditions. Collectively, these kinetic data support a mechanism by which the binding of ATP to an allosteric site on Lon activates the proteolytic site. In this model, the energy derived from the binding of ATP minimally supports peptide cleavage by allowing peptide substrate access to the proteolytic site. However, the kinetics of peptide cleavage are enhanced by the hydrolysis of ATP.
Figures











Similar articles
-
Monitoring the timing of ATP hydrolysis with activation of peptide cleavage in Escherichia coli Lon by transient kinetics.Biochemistry. 2005 Feb 8;44(5):1671-82. doi: 10.1021/bi048618z. Biochemistry. 2005. PMID: 15683251
-
Transient kinetic experiments demonstrate the existence of a unique catalytic enzyme form in the peptide-stimulated ATPase mechanism of Escherichia coli Lon protease.Biochemistry. 2006 Sep 26;45(38):11432-43. doi: 10.1021/bi060809+. Biochemistry. 2006. PMID: 16981703
-
Processive degradation of unstructured protein by Escherichia coli Lon occurs via the slow, sequential delivery of multiple scissile sites followed by rapid and synchronized peptide bond cleavage events.Biochemistry. 2013 Aug 20;52(33):5629-44. doi: 10.1021/bi4008319. Epub 2013 Jul 24. Biochemistry. 2013. PMID: 23822859
-
Recent developments in the mechanistic enzymology of the ATP-dependent Lon protease from Escherichia coli: highlights from kinetic studies.Mol Biosyst. 2006 Oct;2(10):477-83. doi: 10.1039/b609936j. Epub 2006 Aug 24. Mol Biosyst. 2006. PMID: 17216028 Review.
-
Utilization of positional isotope exchange experiments to evaluate reversibility of ATP hydrolysis catalyzed by Escherichia coli Lon protease.Biochem Cell Biol. 2010 Feb;88(1):119-28. doi: 10.1139/o09-117. Biochem Cell Biol. 2010. PMID: 20130685 Review.
Cited by
-
Structural basis for distinct operational modes and protease activation in AAA+ protease Lon.Sci Adv. 2020 May 20;6(21):eaba8404. doi: 10.1126/sciadv.aba8404. eCollection 2020 May. Sci Adv. 2020. PMID: 32490208 Free PMC article.
-
Degrons in protein substrates program the speed and operating efficiency of the AAA+ Lon proteolytic machine.Proc Natl Acad Sci U S A. 2009 Nov 3;106(44):18503-8. doi: 10.1073/pnas.0910392106. Epub 2009 Oct 19. Proc Natl Acad Sci U S A. 2009. PMID: 19841274 Free PMC article.
-
Utilization of synthetic peptides to evaluate the importance of substrate interaction at the proteolytic site of Escherichia coli Lon protease.Biochim Biophys Acta. 2009 Sep;1794(9):1355-63. doi: 10.1016/j.bbapap.2009.02.015. Epub 2009 Mar 11. Biochim Biophys Acta. 2009. PMID: 19285157 Free PMC article.
-
Active-site-directed chemical tools for profiling mitochondrial Lon protease.ACS Chem Biol. 2011 Aug 19;6(8):781-8. doi: 10.1021/cb100408w. Epub 2011 May 6. ACS Chem Biol. 2011. PMID: 21520912 Free PMC article.
-
The peroxisomal Lon protease LonP2 in aging and disease: functions and comparisons with mitochondrial Lon protease LonP1.Biol Rev Camb Philos Soc. 2017 May;92(2):739-753. doi: 10.1111/brv.12253. Epub 2016 Feb 8. Biol Rev Camb Philos Soc. 2017. PMID: 26852705 Free PMC article. Review.
References
-
- Lee I, Berdis AJ. Adenosine triphosphate-dependent degradation of a fluorescent lambda N substrate mimic by Lon protease. Anal Biochem. 2001;291:74–83. - PubMed
-
- Thomas-Wohlever J, Lee I. Kinetic characterization of the peptidase activity of Escherichia coli Lon reveals the mechanistic similarities in ATP-dependent hydrolysis of peptide and protein substrates. Biochemistry. 2002;41:9418–9425. - PubMed
-
- Goff SA, Goldberg AL. Production of abnormal proteins in E. coli stimulates transcription of lon and other heat shock genes. Cell. 1985;41:587–595. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

