Broad antiretroviral activity and resistance profile of the novel human immunodeficiency virus integrase inhibitor elvitegravir (JTK-303/GS-9137)
- PMID: 17977962
- PMCID: PMC2224569
- DOI: 10.1128/JVI.01534-07
Broad antiretroviral activity and resistance profile of the novel human immunodeficiency virus integrase inhibitor elvitegravir (JTK-303/GS-9137)
Abstract
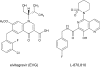
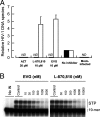
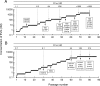
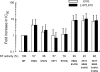
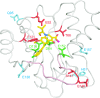
Integrase (IN), an essential enzyme of human immunodeficiency virus (HIV), is an attractive antiretroviral drug target. The antiviral activity and resistance profile in vitro of a novel IN inhibitor, elvitegravir (EVG) (also known as JTK-303/GS-9137), currently being developed for the treatment of HIV-1 infection are described. EVG blocked the integration of HIV-1 cDNA through the inhibition of DNA strand transfer. EVG inhibited the replication of HIV-1, including various subtypes and multiple-drug-resistant clinical isolates, and HIV-2 strains with a 50% effective concentration in the subnanomolar to nanomolar range. EVG-resistant variants were selected in two independent inductions, and a total of 8 amino acid substitutions in the catalytic core domain of IN were observed. Among the observed IN mutations, T66I and E92Q substitutions mainly contributed to EVG resistance. These two primary resistance mutations are located in the active site, and other secondary mutations identified are proximal to these primary mutations. The EVG-selected IN mutations, some of which represent novel IN inhibitor resistance mutations, conferred reduced susceptibility to other IN inhibitors, suggesting that a common mechanism is involved in resistance and potential cross-resistance. The replication capacity of EVG-resistant variants was significantly reduced relative to both wild-type virus and other IN inhibitor-resistant variants selected by L-870,810. EVG and L-870,810 both inhibited the replication of murine leukemia virus and simian immunodeficiency virus, suggesting that IN inhibitors bind to a conformationally conserved region of various retroviral IN enzymes and are an ideal drug for a range of retroviral infections.
Figures

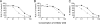
References
-
- Asante-Appiah, E., and A. M. Skalka. 1997. Molecular mechanisms in retrovirus DNA integration. Antivir. Res. 36139-156. - PubMed
-
- Brown, P. O., B. Bowerman, H. E. Varmus, and J. M. Bishop. 1987. Correct integration of retroviral DNA in vitro. Cell 49347-356. - PubMed
-
- Bukrinsky, M. I., N. Sharova, T. L. McDonald, T. Pushkarskaya, W. G. Tarpley, and M. Stevenson. 1993. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. USA 906125-6129. - PMC - PubMed
-
- Butler, S. L., M. S. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7631-634. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

