Effects of partial deletions within the human immunodeficiency virus type 1 V3 loop on coreceptor tropism and sensitivity to entry inhibitors
- PMID: 17977968
- PMCID: PMC2224606
- DOI: 10.1128/JVI.01793-07
Effects of partial deletions within the human immunodeficiency virus type 1 V3 loop on coreceptor tropism and sensitivity to entry inhibitors
Abstract
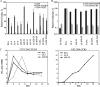
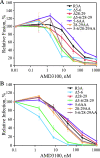
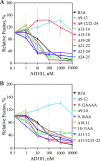
The human immunodeficiency virus type 1 (HIV-1) V3 loop is critical for coreceptor binding and principally determines tropism for the CCR5 and CXCR4 coreceptors. The recent crystallographic resolution of V3 shows that its base is closely associated with the conserved coreceptor binding site on the gp120 core, whereas more distal regions protrude toward the cell surface, likely mediating interactions with coreceptor extracellular loops. However, these V3-coreceptor interactions and the structural basis for CCR5 or CXCR4 specificity are poorly understood. Using the dual-tropic virus HIV-1(R3A), which uses both CCR5 and CXCR4, we sought to identify subdomains within V3 that selectively mediate R5 or X4 tropism. An extensive panel of V3 mutants was evaluated for effects on tropism and sensitivity to coreceptor antagonists. Mutations on either side of the V3 base (residues 3 to 8 and 26 to 33) ablated R5 tropism and made the resulting X4-tropic Envs more sensitive to the CXCR4 inhibitor AMD3100. When mutations were introduced within the V3 stem, only a deletion of residues 9 to 12 on the N-terminal side ablated X4 tropism. Remarkably, this R5-tropic Delta9-12 mutant was completely resistant to several small-molecule inhibitors of CCR5. Envs with mutations in the V3 crown (residues 13 to 20) remained dual tropic. Similar observations were made for a second dual-tropic isolate, HIV-1(89.6). These findings suggest that V3 subdomains can be identified that differentially affect R5 and X4 tropism and modulate sensitivity to CCR5 and CXCR4 inhibitors. These studies provide a novel approach for probing V3-coreceptor interactions and mechanisms by which these interactions can be inhibited.
Figures

References
-
- Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 2721955-1958. - PubMed
-
- Atchison, R. E., J. Gosling, F. S. Monteclaro, C. Franci, L. Digilio, I. F. Charo, and M. A. Goldsmith. 1996. Multiple extracellular elements of CCR5 and HIV-1 entry: dissociation from response to chemokines. Science 2741924-1926. - PubMed
-
- Brelot, A., N. Heveker, M. Montes, and M. Alizon. 2000. Identification of residues of CXCR4 critical for human immunodeficiency virus coreceptor and chemokine receptor activities. J. Biol. Chem. 27523736-23744. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

