fimA genotypes and multilocus sequence types of Porphyromonas gingivalis from patients with periodontitis
- PMID: 17977992
- PMCID: PMC2224254
- DOI: 10.1128/JCM.00986-07
fimA genotypes and multilocus sequence types of Porphyromonas gingivalis from patients with periodontitis
Abstract
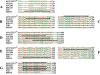
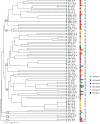
Fimbriae are important virulence factors of pathogenic bacteria, facilitating their attachment to host and bacterial cells. In the periodontal pathogen Porphyromonas gingivalis, the fimA gene is classified into six types (genotypes I, Ib, II, III, IV, and V) on the basis of different nucleotide sequences, with fimA genotypes II and IV being prevalent in isolates from patients with periodontitis. The aims of this study were to examine the distribution of fimA genotypes in a collection of 82 P. gingivalis isolates from adult periodontitis patients of worldwide origin and to investigate the relationship between the fimA genotypes and the sequence types (STs), as determined by multilocus sequence typing (MLST), of the isolates. The fimA gene was amplified by PCR with primer sets specific for each genotype. The STs of all strains were assigned according to the MLST database for P. gingivalis (www.pubmlst.org/pgingivalis). The 82 strains showed extensive genetic diversity and were assigned to 69 STs. Only isolates with closely related STs harbored the same fimA genotype. Twenty-eight (34.1%) strains harbored fimA genotype II, while only the reference strain for fimA genotype V reacted with the primers specific for this genotype. Twenty-one isolates (25.6%) were positive by more than one of the fimA PCR assays; the most frequent combinations were genotypes I, Ib, and II (eight isolates) and genotypes I and II (four isolates). Sequencing of the fimA gene from selected isolates did not support the observed specific fimA genotype combinations, suggesting that the genotyping method used for the major fimbriae in P. gingivalis should be reevaluated.
Figures

Similar articles
-
Distribution of Porphyromonas gingivalis fimA genotypes in isolates from subgingival plaque and blood sample during bacteremia.Biomedica. 2009 Jun;29(2):298-306. Biomedica. 2009. PMID: 20128354
-
Distribution and molecular characterization of Porphyromonas gingivalis carrying a new type of fimA gene.J Clin Microbiol. 2000 May;38(5):1909-14. doi: 10.1128/JCM.38.5.1909-1914.2000. J Clin Microbiol. 2000. PMID: 10790120 Free PMC article.
-
Distribution of Porphyromonas gingivalis strains with fimA genotypes in periodontitis patients.J Clin Microbiol. 1999 May;37(5):1426-30. doi: 10.1128/JCM.37.5.1426-1430.1999. J Clin Microbiol. 1999. PMID: 10203499 Free PMC article.
-
Functional differences of Porphyromonas gingivalis Fimbriae in determining periodontal disease pathogenesis: a literature review.Colomb Med (Cali). 2013 Mar 30;44(1):48-56. eCollection 2013 Jan. Colomb Med (Cali). 2013. PMID: 24892323 Free PMC article. Review.
-
Variations of Porphyromonas gingivalis fimbriae in relation to microbial pathogenesis.J Periodontal Res. 2004 Apr;39(2):136-42. doi: 10.1111/j.1600-0765.2004.00719.x. J Periodontal Res. 2004. PMID: 15009522 Review.
Cited by
-
Genetic diversity in the oral pathogen Porphyromonas gingivalis: molecular mechanisms and biological consequences.Future Microbiol. 2013 May;8(5):607-20. doi: 10.2217/fmb.13.30. Future Microbiol. 2013. PMID: 23642116 Free PMC article. Review.
-
Multilocus sequence analysis of Treponema denticola strains of diverse origin.BMC Microbiol. 2013 Feb 4;13:24. doi: 10.1186/1471-2180-13-24. BMC Microbiol. 2013. PMID: 23379917 Free PMC article.
-
A Tale of Two Fimbriae: How Invasion of Dendritic Cells by Porphyromonas gingivalis Disrupts DC Maturation and Depolarizes the T-Cell-Mediated Immune Response.Pathogens. 2022 Mar 8;11(3):328. doi: 10.3390/pathogens11030328. Pathogens. 2022. PMID: 35335652 Free PMC article. Review.
-
Genetic exchange of fimbrial alleles exemplifies the adaptive virulence strategy of Porphyromonas gingivalis.PLoS One. 2014 Mar 13;9(3):e91696. doi: 10.1371/journal.pone.0091696. eCollection 2014. PLoS One. 2014. PMID: 24626479 Free PMC article.
-
Oral dysbiosis induced by Porphyromonas gingivalis is strain-dependent in mice.J Oral Microbiol. 2020 Oct 12;12(1):1832837. doi: 10.1080/20002297.2020.1832837. J Oral Microbiol. 2020. PMID: 33133418 Free PMC article.
References
-
- Al-Zahrani, M. S., N. Bissada, and E. A. Borawski. 2003. Obesity and periodontal disease in young, middle-aged and older adults. J. Periodontol. 74610-615. - PubMed
-
- Amano, A., M. Kubinowa, I. Nakagawa, S. Akiyama, I. Morisaki, and S. Hamada. 2000. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J. Dent. Res. 791664-1668. - PubMed
-
- Amano, A. 2003. Molecular interaction of Porphyromonas gingivalis with host cells: implication for the microbial pathogenesis of periodontal disease. J. Periodontol. 7490-96. - PubMed
-
- Amano, A., I. Nakagawa, N. Okahashi, and N. Hamada. 2004. Variations of Porphyromonas gingivalis fimbriae in relation to microbial pathogenesis. J. Periodontal Res. 39136-142. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

