Genome-wide analyses of human perisylvian cerebral cortical patterning
- PMID: 17978184
- PMCID: PMC2077018
- DOI: 10.1073/pnas.0706128104
Genome-wide analyses of human perisylvian cerebral cortical patterning
Abstract
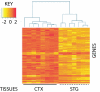
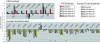
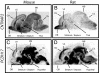
Despite the well established role of the frontal and posterior perisylvian cortices in many facets of human-cognitive specializations, including language, little is known about the developmental patterning of these regions in the human brain. We performed a genome-wide analysis of human cerebral patterning during midgestation, a critical epoch in cortical regionalization. A total of 345 genes were identified as differentially expressed between superior temporal gyrus (STG) and the remaining cerebral cortex. Gene ontology categories representing transcription factors were enriched in STG, whereas cell-adhesion and extracellular matrix molecules were enriched in the other cortical regions. Quantitative RT-PCR or in situ hybridization was performed to validate differential expression in a subset of 32 genes, most of which were confirmed. LIM domain-binding 1 (LDB1), which we show to be enriched in the STG, is a recently identified interactor of LIM domain only 4 (LMO4), a gene known to be involved in the asymmetric pattering of the perisylvian region in the developing human brain. Protocadherin 17 (PCDH17), a neuronal cell adhesion molecule, was highly enriched in focal regions of the human prefrontal cortex. Contactin associated protein-like 2 (CNTNAP2), in which mutations are known to cause autism, epilepsy, and language delay, showed a remarkable pattern of anterior-enriched cortical expression in human that was not observed in mouse or rat. These data highlight the importance of expression analysis of human brain and the utility of cross-species comparisons of gene expression. Genes identified here provide a foundation for understanding molecular aspects of human-cognitive specializations and the disorders that disrupt them.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Hauser MD, Chomsky N, Fitch WT. Science. 2002;298:1569–1579. - PubMed
-
- Geschwind N. Science. 1970;170:940–944. - PubMed
-
- Galaburda AM, LeMay M, Kemper TL, Geschwind N. Science. 1978;199:852–856. - PubMed
-
- Lieberman P. Am J Phys Anthropol. 2002;35(Suppl):36–62. - PubMed
-
- Baynes K, Eliassen JC, Lutsep HL, Gazzaniga MS. Science. 1998;280:902–905. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

