Extracting gene expression patterns and identifying co-expressed genes from microarray data reveals biologically responsive processes
- PMID: 17980031
- PMCID: PMC2194742
- DOI: 10.1186/1471-2105-8-427
Extracting gene expression patterns and identifying co-expressed genes from microarray data reveals biologically responsive processes
Abstract
Background: A common observation in the analysis of gene expression data is that many genes display similarity in their expression patterns and therefore appear to be co-regulated. However, the variation associated with microarray data and the complexity of the experimental designs make the acquisition of co-expressed genes a challenge. We developed a novel method for Extracting microarray gene expression Patterns and Identifying co-expressed Genes, designated as EPIG. The approach utilizes the underlying structure of gene expression data to extract patterns and identify co-expressed genes that are responsive to experimental conditions.
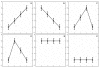
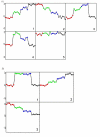
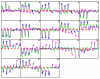
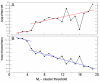
Results: Through evaluation of the correlations among profiles, the magnitude of variation in gene expression profiles, and profile signal-to-noise ratio's, EPIG extracts a set of patterns representing co-expressed genes. The method is shown to work well with a simulated data set and microarray data obtained from time-series studies of dauer recovery and L1 starvation in C. elegans and after ultraviolet (UV) or ionizing radiation (IR)-induced DNA damage in diploid human fibroblasts. With the simulated data set, EPIG extracted the appropriate number of patterns which were more stable and homogeneous than the set of patterns that were determined using the CLICK or CAST clustering algorithms. However, CLICK performed better than EPIG and CAST with respect to the average correlation between clusters/patterns of the simulated data. With real biological data, EPIG extracted more dauer-specific patterns than CLICK. Furthermore, analysis of the IR/UV data revealed 18 unique patterns and 2661 genes out of approximately 17,000 that were identified as significantly expressed and categorized to the patterns by EPIG. The time-dependent patterns displayed similar and dissimilar responses between IR and UV treatments. Gene Ontology analysis applied to each pattern-related subset of co-expressed genes revealed underlying biological processes affected by IR- and/or UV- induced DNA damage.
Conclusion: EPIG competed with CLICK and performed better than CAST in extracting patterns from simulated data. EPIG extracted more biological informative patterns and co-expressed genes from both C. elegans and IR/UV-treated human fibroblasts. Using Gene Ontology analysis of the genes in the patterns extracted by EPIG, several key biological categories related to p53-dependent cell cycle control were revealed from the IR/UV data. Among them were mitotic cell cycle, DNA replication, DNA repair, cell cycle checkpoint, and G0-like status transition. EPIG can be applied to data sets from a variety of experimental designs.
Figures


References
-
- Eisen MB, Brown PO. DNA arrays for analysis of gene expression. Methods Enzymol. 1999;303:179–205. - PubMed
-
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci U S A. 1999;96:2907–2912. doi: 10.1073/pnas.96.6.2907. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

