S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase
- PMID: 17980632
- PMCID: PMC4979535
- DOI: 10.1016/j.immuni.2007.09.007
S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase
Abstract
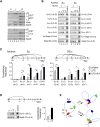
Molecular mechanisms underlying synapsis of activation-induced deaminase (AID)-targeted S regions during class switch recombination (CSR) are poorly understood. By using chromosome conformation capture techniques, we found that in B cells, the Emicro and 3'Ealpha enhancers were in close spatial proximity, forming a unique chromosomal loop configuration. B cell activation led to recruitment of the germline transcript (GLT) promoters to the Emicro:3'Ealpha complex in a cytokine-dependent fashion. This structure facilitated S-S synapsis because Smicro was proximal to Emicro and a downstream S region was corecruited with the targeted GLT promoter to Emicro:3'Ealpha. We propose that GLT promoter association with the Emicro:3'Ealpha complex creates an architectural scaffolding that promotes S-S synapsis during CSR and that these interactions are stabilized by AID. Thus, the S-S synaptosome is formed as a result of the self-organizing transcription system that regulates GLT expression and may serve to guard against spurious chromosomal translocations.
Conflict of interest statement
The authors declare that they have no competing financial interests.
Figures


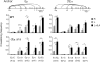

Similar articles
-
Loop extrusion promotes an alternate pathway for isotype switching.Cell Rep. 2021 Nov 23;37(8):110059. doi: 10.1016/j.celrep.2021.110059. Cell Rep. 2021. PMID: 34818547 Free PMC article.
-
53BP1 Contributes to Igh Locus Chromatin Topology during Class Switch Recombination.J Immunol. 2017 Mar 15;198(6):2434-2444. doi: 10.4049/jimmunol.1601947. Epub 2017 Feb 3. J Immunol. 2017. PMID: 28159901 Free PMC article.
-
Translocation of an antibody transgene requires AID and occurs by interchromosomal switching to all switch regions except the mu switch region.Eur J Immunol. 2011 May;41(5):1456-64. doi: 10.1002/eji.201041077. Epub 2011 Apr 11. Eur J Immunol. 2011. PMID: 21469111 Free PMC article.
-
Current insights into the mechanism of mammalian immunoglobulin class switch recombination.Crit Rev Biochem Mol Biol. 2019 Aug;54(4):333-351. doi: 10.1080/10409238.2019.1659227. Epub 2019 Sep 11. Crit Rev Biochem Mol Biol. 2019. PMID: 31509023 Free PMC article. Review.
-
Regulating infidelity: RNA-mediated recruitment of AID to DNA during class switch recombination.Eur J Immunol. 2016 Mar;46(3):523-30. doi: 10.1002/eji.201545809. Epub 2016 Feb 22. Eur J Immunol. 2016. PMID: 26799454 Free PMC article. Review.
Cited by
-
Mice lacking Sμ tandem repeats maintain RNA polymerase patterns but exhibit histone modification pattern shifts linked to class switch site locations.Mol Immunol. 2012 Aug;52(1):1-8. doi: 10.1016/j.molimm.2012.04.006. Epub 2012 May 11. Mol Immunol. 2012. PMID: 22580346 Free PMC article.
-
Developmental regulation of DNA cytosine methylation at the immunoglobulin heavy chain constant locus.PLoS Genet. 2019 Feb 19;15(2):e1007930. doi: 10.1371/journal.pgen.1007930. eCollection 2019 Feb. PLoS Genet. 2019. PMID: 30779742 Free PMC article.
-
Generating and repairing genetically programmed DNA breaks during immunoglobulin class switch recombination.F1000Res. 2018 Apr 13;7:458. doi: 10.12688/f1000research.13247.1. eCollection 2018. F1000Res. 2018. PMID: 29744038 Free PMC article. Review.
-
Complex relationship between mismatch repair proteins and MBD4 during immunoglobulin class switch recombination.PLoS One. 2013 Oct 29;8(10):e78370. doi: 10.1371/journal.pone.0078370. eCollection 2013. PLoS One. 2013. PMID: 24205214 Free PMC article.
-
BRD2 promotes antibody class switch recombination by facilitating DNA repair in collaboration with NIPBL.Nucleic Acids Res. 2024 May 8;52(8):4422-4439. doi: 10.1093/nar/gkae204. Nucleic Acids Res. 2024. PMID: 38567724 Free PMC article.
References
-
- Aten JA, Stap J, Krawczyk PM, van Oven CH, Hoebe RA, Essers J, Kanaar R. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004;303:92–95. - PubMed
-
- Barreto V, Reina-San-Martin B, Ramiro AR, McBride KM, Nussenzweig MC. C-terminal deletion of AID uncouples class switch recombination from somatic hypermutation and gene conversion. Mol Cell. 2003;12:501–508. - PubMed
-
- Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–626. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

