Functional specificity among ribosomal proteins regulates gene expression
- PMID: 17981122
- PMCID: PMC2443060
- DOI: 10.1016/j.cell.2007.08.037
Functional specificity among ribosomal proteins regulates gene expression
Abstract
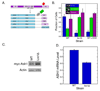
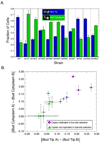
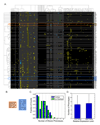
Duplicated genes escape gene loss by conferring a dosage benefit or evolving diverged functions. The yeast Saccharomyces cerevisiae contains many duplicated genes encoding ribosomal proteins. Prior studies have suggested that these duplicated proteins are functionally redundant and affect cellular processes in proportion to their expression. In contrast, through studies of ASH1 mRNA in yeast, we demonstrate paralog-specific requirements for the translation of localized mRNAs. Intriguingly, these paralog-specific effects are limited to a distinct subset of duplicated ribosomal proteins. Moreover, transcriptional and phenotypic profiling of cells lacking specific ribosomal proteins reveals differences between the functional roles of ribosomal protein paralogs that extend beyond effects on mRNA localization. Finally, we show that ribosomal protein paralogs exhibit differential requirements for assembly and localization. Together, our data indicate complex specialization of ribosomal proteins for specific cellular processes and support the existence of a ribosomal code.
Figures
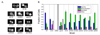
Comment in
-
Yeast ribosomes: variety is the spice of life.Cell. 2007 Nov 2;131(3):450-1. doi: 10.1016/j.cell.2007.10.028. Cell. 2007. PMID: 17981113
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

