The PAR proteins: fundamental players in animal cell polarization
- PMID: 17981131
- PMCID: PMC2964935
- DOI: 10.1016/j.devcel.2007.10.007
The PAR proteins: fundamental players in animal cell polarization
Abstract
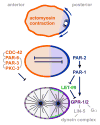
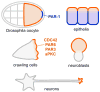
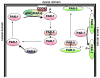
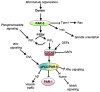
The par genes were discovered in genetic screens for regulators of cytoplasmic partitioning in the early embryo of C. elegans, and encode six different proteins required for asymmetric cell division by the worm zygote. Some of the PAR proteins are localized asymmetrically and form physical complexes with one another. Strikingly, the PAR proteins have been found to regulate cell polarization in many different contexts in diverse animals, suggesting they form part of an ancient and fundamental mechanism for cell polarization. Although the picture of how the PAR proteins function remains incomplete, cell biology and biochemistry are beginning to explain how PAR proteins polarize cells.
Figures

References
-
- Affolter M, Weijer CJ. Signaling to cytoskeletal dynamics during chemotaxis. Dev Cell. 2005;9:19–34. - PubMed
-
- Alessi DR, Sakamoto K, Bayascas JR. LKB1-dependent signaling pathways. Annu Rev Biochem. 2006;75:137–163. - PubMed
-
- Baas AF, Smit L, Clevers H. LKB1 tumor suppressor protein: PARtaker in cell polarity. Trends Cell Biol. 2004;14:312–319. - PubMed
-
- Barnes AP, Lilley BN, Pan YA, Plummer LJ, Powell AW, Raines AN, Sanes JR, Polleux F. LKB1 and SAD kinases define a pathway required for the polarization of cortical neurons. Cell. 2007;129:549–563. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

