Insights into COPII coat nucleation from the structure of Sec23.Sar1 complexed with the active fragment of Sec31
- PMID: 17981133
- PMCID: PMC2686382
- DOI: 10.1016/j.devcel.2007.10.006
Insights into COPII coat nucleation from the structure of Sec23.Sar1 complexed with the active fragment of Sec31
Abstract
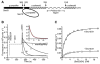
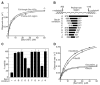
The COPII vesicular coat forms on the endoplasmic reticulum from Sar1-GTP, Sec23/24 and Sec13/31 protein subunits. Here, we define the interaction between Sec23/24.Sar1 and Sec13/31, involving a 40 residue Sec31 fragment. In the crystal structure of the ternary complex, Sec31 binds as an extended polypeptide across a composite surface of the Sec23 and Sar1-GTP molecules, explaining the stepwise character of Sec23/24.Sar1 and Sec13/31 recruitment to the membrane. The Sec31 fragment stimulates GAP activity of Sec23/24, and a convergence of Sec31 and Sec23 residues at the Sar1 GTPase active site explains how GTP hydrolysis is triggered leading to COPII coat disassembly. The Sec31 active fragment is accommodated in a binding groove supported in part by Sec23 residue Phe380. Substitution of the corresponding residue F382L in human Sec23A causes cranio-lenticulo-sutural dysplasia, and we suggest that this mutation disrupts the nucleation of COPII coat proteins at endoplasmic reticulum exit sites.
Figures



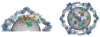
References
-
- Ahmadian MR, Hoffman U, Goody RS, Wittinghofer A. Individual rate constants for the interaction of Ras proteins with GTPase-activating proteins determined by fluorescence spectroscopy. Biochemistry. 1997;36:4535–4541. - PubMed
-
- Antonny B, Beraud-Dufour S, Chardin P, Chabre M. N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry. 1997;36:4675–4684. - PubMed
-
- Antonny B, Madden D, Hamamoto S, Orci L, Schekman R. Dynamics of the COPII coat with GTP and stable analogues. Nat Cell Biol. 2001;3:531–537. - PubMed
-
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. - PubMed
-
- Bi X, Corpina RA, Goldberg J. Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature. 2002;419:271–277. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

