SNF1/AMPK pathways in yeast
- PMID: 17981722
- PMCID: PMC2685184
- DOI: 10.2741/2854
SNF1/AMPK pathways in yeast
Abstract
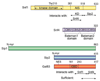
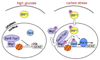
The SNF1/AMPK family of protein kinases is highly conserved in eukaryotes and is required for energy homeostasis in mammals, plants, and fungi. SNF1 protein kinase was initially identified by genetic analysis in the budding yeast Saccharomyces cerevisiae. SNF1 is required primarily for the adaptation of yeast cells to glucose limitation and for growth on carbon sources that are less preferred than glucose, but is also involved in responses to other environmental stresses. SNF1 regulates transcription of a large set of genes, modifies the activity of metabolic enzymes, and controls various nutrient-responsive cellular developmental processes. Like AMPK, SNF1 protein kinase is heterotrimeric. It is phosphorylated and activated by the upstream kinases Sak1, Tos3, and Elm1 and is inactivated by the Reg1-Glc7 protein phosphatase 1. Further regulation of SNF1 is achieved through autoinhibition and through control of its subcellular localization. Here we review the current understanding of SNF1 protein kinase pathways in Saccharomyces cerevisiae and other yeasts.
Figures


References
-
- Carlson M, Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted and intracellular forms of yeast invertase. Cell. 1982;28:145–154. - PubMed
-
- Ciriacy M. Isolation and characterization of yeast mutants defective in intermediary carbon metabolism and in carbon catabolite derepression. Mol. Gen. Genet. 1977;154:213–220. - PubMed
-
- Zimmermann FK, Scheel I. Mutants of Saccharomyces cerevisiae resistant to carbon catabolite repression. Mol. Gen. Genet. 1977;154:75–82. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
