Packaging the genome: the structure of mitotic chromosomes
- PMID: 17981824
- PMCID: PMC3943392
- DOI: 10.1093/jb/mvm214
Packaging the genome: the structure of mitotic chromosomes
Abstract
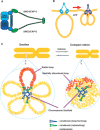
Mitotic chromosomes are essential structures for the faithful transmission of duplicated genomic DNA into two daughter cells during cell division. Although more than 100 years have passed since chromosomes were first observed, it remains unclear how a long string of genomic DNA is packaged into compact mitotic chromosomes. Although the classical view is that human chromosomes consist of radial 30 nm chromatin loops that are somehow tethered centrally by scaffold proteins, called condensins, cryo-electron microscopy observation of frozen hydrated native chromosomes reveals a homogeneous, grainy texture and neither higher-order nor periodic structures including 30 nm chromatin fibres were observed. As a compromise to fill this huge gap, we propose a model in which the radial chromatin loop structures in the classic view are folded irregularly toward the chromosome centre with the increase in intracellular cations during mitosis. Consequently, compact native chromosomes are made up primarily of irregular chromatin networks cross-linked by self-assembled condensins forming the chromosome scaffold.
Figures


References
-
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. - PubMed
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

