Regulatory mechanisms for 3'-end alternative splicing and polyadenylation of the Glial Fibrillary Acidic Protein, GFAP, transcript
- PMID: 17981838
- PMCID: PMC2190720
- DOI: 10.1093/nar/gkm931
Regulatory mechanisms for 3'-end alternative splicing and polyadenylation of the Glial Fibrillary Acidic Protein, GFAP, transcript
Abstract
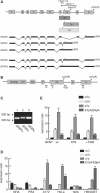
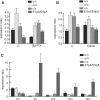
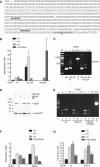
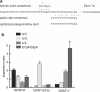
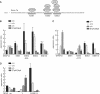
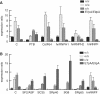
The glial fibrillary acidic protein, GFAP, forms the intermediate cytoskeleton in cells of the glial lineage. Besides the common GFAP alpha transcript, the GFAP epsilon and GFAP kappa transcripts are generated by alternative mRNA 3'-end processing. Here we use a GFAP minigene to characterize molecular mechanisms participating in alternative GFAP expression. Usage of a polyadenylation signal within the alternatively spliced exon 7a is essential to generate the GFAP kappa and GFAP kappa transcripts. The GFAP kappa mRNA is distinct from GFAP epsilon mRNA given that it also includes intron 7a. Polyadenylation at the exon 7a site is stimulated by the upstream splice site. Moreover, exon 7a splice enhancer motifs supported both exon 7a splicing and polyadenylation. SR proteins increased the usage of the exon 7a polyadenylation signal but not the exon 7a splicing, whereas the polypyrimidine tract binding (PTB) protein enhanced both exon 7a polyadenylation and exon 7a splicing. Finally, increasing transcription by the VP16 trans-activator did not affect the frequency of use of the exon 7a polyadenylation signal whereas the exon 7a splicing frequency was decreased. Our data suggest a model with the selection of the exon 7a polyadenylation site being the essential and primary event for regulating GFAP alternative processing.
Figures
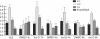

References
-
- Eng LF, Ghirnikar RS, Lee YL. Glial fibrillary acidic protein: GFAP-thirty-one years (1969–2000) Neurochem Res. 2000;25:1439–1451. - PubMed
-
- Parry DA, Steinert PM. Intermediate filament structure. Curr. Opin. Cell. Biol. 1992;4:94–98. - PubMed
-
- Zelenika D, Grima B, Brenner M, Pessac B. A novel glial fibrillary acidic protein mRNA lacking exon 1. Brain Res. Mol. Brain Res. 1995;30:251–258. - PubMed
-
- Condorelli DF, Nicoletti VG, Barresi V, Conticello SG, Caruso A, Tendi EA, Giuffrida Stella AM. Structural features of the rat GFAP gene and identification of a novel alternative transcript. J. Neurosci. Res. 1999;56:219–228. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

