Packaging of a polymer by a viral capsid: the interplay between polymer length and capsid size
- PMID: 17981893
- PMCID: PMC2212672
- DOI: 10.1529/biophysj.107.117473
Packaging of a polymer by a viral capsid: the interplay between polymer length and capsid size
Abstract
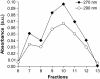
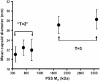
We report a study of the in vitro self-assembly of virus-like particles formed by the capsid protein of cowpea chlorotic mottle virus and the anionic polymer poly(styrene sulfonate) (PSS) for five molecular masses ranging from 400 kDa to 3.4 MDa. The goal is to explore the effect on capsid size of the competition between the preferred curvature of the protein and the molecular mass of the packaged cargo. The capsid size distribution for each polymer was unimodal, but two distinct sizes were observed: 22 nm for the lower molecular masses, jumping to 27 nm at a molecular mass of 2 MDa. A model is provided for the formation of the virus-like particles that accounts for both the PSS and capsid protein self-interactions and the interactions between the protein and PSS. Our study suggests that the size of the encapsidated polymer cargo is the deciding factor for the selection of one distinct capsid size from several possible sizes with the same inherent symmetry.
Figures



References
-
- Crick, F. H. C., and J. D. Watson. 1956. Structure of small viruses. Nature. 177:473–475. - PubMed
-
- Johnson, J. E., and R. R. Rueckert. 1997. Packaging and release of the viral genome. In Structural Biology of Viruses. W. Chiu, R. M. Burnett, and R. L. Garcea, editors. Oxford University Press, New York. 269–287.
-
- Smith, D. E., S. J. Tans, S. B. Smith, S. Grimes, D. L. Anderson, and C. Bustamante. 2001. The bacteriophage phi 29 portal motor can package DNA against a large internal force. Nature. 413:748–752. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

