Theoretical calculations of the catalytic triad in short-chain alcohol dehydrogenases/reductases
- PMID: 17981907
- PMCID: PMC2212670
- DOI: 10.1529/biophysj.107.111096
Theoretical calculations of the catalytic triad in short-chain alcohol dehydrogenases/reductases
Abstract
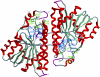
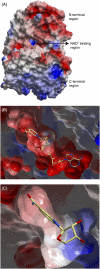
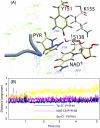
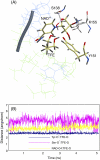
Three highly conserved active site residues (Ser, Tyr, and Lys) of the family of short-chain alcohol dehydrogenases/reductases (SDRs) were demonstrated to be essential for catalytic activity and have been denoted the catalytic triad of SDRs. In this study computational methods were adopted to study the ionization properties of these amino acids in SDRs from Drosophila melanogaster and Drosophila lebanonensis. Three enzyme models, with different ionization scenarios of the catalytic triad that might be possible when inhibitors bind to the enzyme cofactor complex, were constructed. The binding of the two alcohol competitive inhibitors were studied using automatic docking by the Internal Coordinate Mechanics program, molecular dynamic (MD) simulations with the AMBER program package, calculation of the free energy of ligand binding by the linear interaction energy method, and the hydropathic interactions force field. The calculations indicated that deprotonated Tyr acts as a strong base in the binary enzyme-NAD(+) complex. Molecular dynamic simulations for 5 ns confirmed that deprotonated Tyr is essential for anchoring and orientating the inhibitors at the active site, which might be a general trend for the family of SDRs. The findings here have implications for the development of therapeutically important SDR inhibitors.
Figures

Similar articles
-
The catalytic triad in Drosophila alcohol dehydrogenase: pH, temperature and molecular modelling studies.J Mol Biol. 1999 Nov 26;294(2):601-16. doi: 10.1006/jmbi.1999.3235. J Mol Biol. 1999. PMID: 10610783
-
Enzymic and structural studies on Drosophila alcohol dehydrogenase and other short-chain dehydrogenases/reductases.J Mol Evol. 2001 May;52(5):457-66. doi: 10.1007/s002390010175. J Mol Evol. 2001. PMID: 11443349
-
An intact eight-membered water chain in drosophilid alcohol dehydrogenases is essential for optimal enzyme activity.FEBS J. 2012 Aug;279(16):2940-56. doi: 10.1111/j.1742-4658.2012.08675.x. Epub 2012 Jul 19. FEBS J. 2012. PMID: 22741949
-
Medium- and short-chain dehydrogenase/reductase gene and protein families : the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes.Cell Mol Life Sci. 2008 Dec;65(24):3895-906. doi: 10.1007/s00018-008-8588-y. Cell Mol Life Sci. 2008. PMID: 19011750 Free PMC article. Review.
-
Structure and function of retinol dehydrogenases of the short chain dehydrogenase/reductase family.Mol Aspects Med. 2003 Dec;24(6):403-9. doi: 10.1016/s0098-2997(03)00036-0. Mol Aspects Med. 2003. PMID: 14585311 Review.
Cited by
-
Applying internal coordinate mechanics to model the interactions between 8R-lipoxygenase and its substrate.BMC Bioinformatics. 2010 Oct 7;11 Suppl 6(Suppl 6):S2. doi: 10.1186/1471-2105-11-S6-S2. BMC Bioinformatics. 2010. PMID: 20946603 Free PMC article.
-
The short-chain oxidoreductase Q9HYA2 from Pseudomonas aeruginosa PAO1 contains an atypical catalytic center.Protein Sci. 2010 May;19(5):1097-103. doi: 10.1002/pro.384. Protein Sci. 2010. PMID: 20340135 Free PMC article.
-
DFT-based prediction of reactivity of short-chain alcohol dehydrogenase.J Comput Aided Mol Des. 2017 Jun;31(6):587-602. doi: 10.1007/s10822-017-0026-5. Epub 2017 May 26. J Comput Aided Mol Des. 2017. PMID: 28550607 Free PMC article.
-
The enzymology of the human prostanoid pathway.Mol Biol Rep. 2020 Jun;47(6):4569-4586. doi: 10.1007/s11033-020-05526-z. Epub 2020 May 19. Mol Biol Rep. 2020. PMID: 32430846 Review.
-
Selective oxidation of alkyl and aryl glyceryl monoethers catalysed by an engineered and immobilised glycerol dehydrogenase.Chem Sci. 2020 Oct 5;11(44):12009-12020. doi: 10.1039/d0sc04471g. Chem Sci. 2020. PMID: 34123216 Free PMC article.
References
-
- Jornvall, H., B. Persson, M. Krook, S. Atrian, R. Gonzalez-Duarte, J. Jeffery, and D. Ghosh. 1995. Short-chain dehydrogenases/reductases (SDR). Biochemistry. 34:6003–6013. - PubMed
-
- Jornvall, H., J. O. Hoog, and B. Persson. 1999. SDR and MDR: completed genome sequences show these protein families to be large, of old origin, and of complex nature. FEBS Lett. 445:261–264. - PubMed
-
- Jornvall, H., J. O. Hoog, B. Persson, and X. Pares. 2000. Pharmacogenetics of the alcohol dehydrogenase system. Pharmacology. 61:184–191. - PubMed
-
- Hoffmann, F., and E. Maser. 2007. Carbonyl reductases and pluripotent hydroxysteroid dehydrogenases of the short-chain dehydrogenase/reductase superfamily. Drug Metab. Rev. 39:87–144. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

